Supplemental Figure 1. The lateral root number in …...2015/05/22 · Supplemental Data. Yang et...
Transcript of Supplemental Figure 1. The lateral root number in …...2015/05/22 · Supplemental Data. Yang et...

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
1
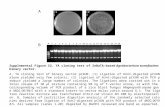
Supplemental Figure 1. The lateral root number in WT, brm-1, brm-3, and
brm-5 roots at 10 DAG.
Supplemental Figure 2. The expression of CycB1;1 and CycB1;3 in 3 DAG WT,
brm-3 and brm-5 roots. Bar=50 μm.

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
2
Supplemental Figure 3. Cellular organization of WT (Col-0), brm-3, brm-5 and
brm-1 root tips at 3 DAG using propidium iodide staining (A) and the length of
4th columella cell of the WT (Col-0), brm-3 and brm-5 root tips at 3 DAG (B).
The QC (white triangle) and columella cell areas (white asterisk) are indicated.
Bars = 20 μm. The 4th columella cell of brm-1 root tips could not be identified.
Data shown are means ± SD (n =20).

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
3
Supplemental Figure 4. The expression of PIN2 in WT (Col-0) and brm-3 root
tips at 3 DAG (A) and the expression of root development related genes in WT
(Col-0) and brm-1 5-day-old seedlings. Bars = 20 μm.

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
4
Supplemental Figure 5. The expression pattern of SHR and SCR in WT and
brm-3 (A, C) and the phenotype of shr-1 brm-3 (B), scr-1 brm-3 (D), shr-1
brm-1 (E) and scr-1 brm-1 (F) double mutants. Bars = 20 μm (A, C) and 1 cm (B,
D, E, F).

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
5
Supplemental Figure 6. The phenotype of plt1-4 plt2-2 brm-3 (A) and plt1-4
plt2-2 brm-1 (B) triple mutants at 7 DAG. Bars = 1 cm.
Supplemental Figure 7. The phenotype of Col-0 and BRMpro:BRM:GFP/brm-1

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
6
seedlings at 10 DAG . Bars = 0.5 cm.
Supplemental Figure 8. ChIP-qPCR analysis of BRM targeting to PIN3, PIN4
and PIN7 (A) and H3K27me3 levels of PIN3, PIN4 and PIN7 loci in brm-3
mutant roots (B). Data are mean values ± standard deviation of three
replicates. Similar results were obtained for at least two additional independent
experiments. Asterisks denote Student’s t- test significant difference between
Col-0 and BRMpro: BRM:GFP/Col-0 roots (p<0.05).

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
7
Supplemental Figure 9. BRM does not target to PLT1 and PLT2 directly.
Supplemental Figure 10. The normal expression of QC25 and QC46 in WT

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
8
(Col-0) and brm-3 root tips at 3 DAG. Bars = 25 μm.
Supplemental Table 1. Primers used for RT-qPCR
Primer
name
Forward Reverse
PLT1 AGTTCGTGGCTGCCATTAGAAGG ATCTTCCATGTTGGTGATGCCTTG
PLT2 GAAGATGGCAAGCAAGGATCGG GCTTCTTCCTCCGTGCTGAATG
PIN1
GGCATGGCTATGTTCAGTCTTGG
G
ACGGCAGGTCCAACGACAAATC
PIN2
TCACGACAACCTCGCTACTAAAG
C
TGCCCATGTAAGGTGACTTTCCC
PIN3 AAGGCGGAAGATCTGACCAAGG TGCTGGATGAGCTACAGCTTTG
PIN4 ACAACGTGGCAACGGAACAATC GCCGATATCATCACCACCACTC
PIN7 CGTGTGGCCATTGTTCAAGCTG
CCCTGTACTCAAGATTGCGGGAT
G
CYB1;1 CACTATTTGGCTGAGTTAGG ATCTTGCTGCGTAGATTGC
CYB1;3 TACTCGTAACTTTCGTGCTCAG GAACCGCTCTTACAACTTCTTG
UBQ10 TCGACCCTTCACTTGGTGTTGC
GGGTGATGGTCTTTCCGGTCAAA
G
Supplemental Table 2. Primers used for ChIP-qPCR
Primer
name
Forward Reverse
Length of
amplicon

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
9
PLT1-P1 gaggacgtgagttcccataatc gcaaattaattatcactcacaatg 132 bp
PLT1-P2 AGGAGAGGTTCCAAAAGTGGCC ATTGGACGCTAGGCATCAAG 131 bp
PLT1-P3 ttccacttaattaggttttgtc ttatacgattcgatgcactcac 150 bp
PLT1-P4 TGGCTGCCATTAGAAGgttt GATACATCGAAGCGCCTCTC 140 bp
PLT1-P5 GGCTCCACCTCATCAAGACT AAGAGGAGGAATCGTGAGCA 133 bp
PLT1-P6 tcaatttagagggggccttt cccataaaaccctaaccctca 120 bp
PLT2-P1 taaacgagggacgtgagtt cgcttccaactagaaaaagtc 130 bp
PLT2-P2 ccaagaaaagggaaATGAA GGTTTTGAAAAGGGTTGTCG 148 bp
PLT2-P3 CAGAAGTCGCCACTGTGAAA CCAAAAGTTTCCAAAGTCCGT 124 bp
PLT2-P4 tcggactatgcactcagGTG tcatgaaaatttgaaataacttacCG 130 bp
PLT2-P5 AACGGCTCAAAGAAGCTCAA AGAGCTGGAAGCAACAGAGC 122 bp
PLT2-P6 acaagtattaggatcggctgga ccctcgaggactctgataacc 120 bp
PIN1-P1 gggataaattgatcaaccgaat ggtggagctttggggatagt 140 bp
PIN1-P2 ttccctcttcaccacttctctc ACGGAACCATAGCCGTCATA 128 bp
PIN1-P3 GAACCCAGGGATGTTTTCG GCACTTGAGCTCCACACAAA 135 bp
PIN1-P4 CCTCCATGTTGCCATTATCC AAGCTGCctgtttgttgtca 125 bp
PIN1-P5 tgccctttttgttgaaaacc aaggtaaaacttgctgagctcct 127 bp
PIN2-P1 ccaatcttccttatcccaaga ggtccataatttttcgaagtgc 137 bp
PIN2-P2 tctctcgccggaaaaagtaa GTGTGAATATCCCCCACCAC 132 bp
PIN2-P3 TCTCACCGGCGTAGAGATTT GCCGCCGTAGCTATTAGTGT 148 bp
PIN2-P4 ttaaatagatataagaatcgatt aattattattattttgttaaaaga 146 bp
PIN2-P5 ttgcaaataaaaggcgatacg attaatgcgacattgcatgg 128 bp

Supplemental Data. Yang et al. (2015). Plant Cell 10.1105/tpc.15.00091
10
PIN3-P1 aaaatcaagcacatgaatgtcac tcgactgattgaaacggaca 122 bp
PIN3-P2 cactgctcccaagtcctctc AGGATCATGGCCACGTAGAG 147 bp
PIN3-P3 AGGTCCAACTCCTCGACCTT GACGCTTTTATTGGCGGTAG 154 bp
PIN3-P4 TGGACTTATTTGGGCTCTCG GATTTTGGGCATTGCCACGTG 135 bp
PIN3-P5 acgcagataaggaggagcaa cccgaacctaatcaaagaaca 153 bp
PIN4-P1 gagttttctggagggacgtg cgtagtattcgccaaagttcc 129 bp
PIN4-P2 tcctcttcaccgaatccttg cggtgggttttggagtttag 144 bp
PIN4-P3 ctgagaaaaccaaaaatcatgaatac tcctcttcaccgaatccttg 120 bp
PIN4-P4 tgaccctctgacccagacat ggatttgagaaaattgacagca 127 bp
PIN4-P5 TCTTGGCCTTTGAattggtc cttccactctttcccacgaa 121 bp
PIN7-P1 aagttttggaccggcattta caatgaattttgtgaatccttga 140 bp
PIN7-P2 tgttaaaagcttccgtcatca AGGACGGTGTAGAGGTCGTG 140 bp
PIN7-P3 CATACTCAAAATGGTGAAAAC ATAGTCCCCGTTCATCGGAC 125 bp
PIN7-P4 ggtgggatgacatagctcgt aacaagtgctgaatgatgcaa 153 bp
PIN7-P5 tctgcttggctgaagaggat ggatcctcaaatcaaatcttgg 125 bp
TA3 ctgcgtggaagtctgtcaaa ctatgccacagggcagttt 106bp
TUB2
ACAAACACAGAGAGGAGTGAGC
A
ACGCATCTTCGGTTGGATGAG
TG
150 bp