Supplemental Figure 1 - Cancer Discovery · Supplemental Figure 6 0 1 Haplotype ratio D:R Chr. 16...
Transcript of Supplemental Figure 1 - Cancer Discovery · Supplemental Figure 6 0 1 Haplotype ratio D:R Chr. 16...

TCGA-06-0214
TCGA-06-5415
TCGA-14-2554
TCGA-26-5132
EGFR vIII var. 1
EGFR vIII var. 2
EGFR vIII var. 3
EGFR C-ter deletion
EGFR vIII var. 1
EGFR vIII var. 2
EGFR vIII var. 1
Δ13-15
EGFR vIII var. 2
EGFR vIII var. 3
EGFR vIII var. 1
EGFR vIII var. 2
EGFR vIII var. 3
A
B
C
D
Supplemental Figure 1

Number of read pairs (million)10
010
110
20
0.5
1
Different blood normal libraries
Bulk primary tumor libraries
Down-sampled bulk tumor libraries
Selected single-cell WGA libraries
Poor WGA libraries
0.5
1
100
101
102
Number of read pairs (million)
KS
met
ric (
~60
Kb
bins
)K
S m
etric
(~
57 K
b bi
ns)
0 0.5 10
0.5
1
KS metric
% o
f pile
up r
eads
rem
oved
Down-sampled bulk tumor librariesSingle-cell WGA libraries
A
C
BBT340
BT325
0
Supplemental Figure 2

7: 54,560,250--55,510,789
p11.1
A
p12.1 p11.2
BT325
EGFR
p12.1 p11.1
7: 54,861,307--55,396,737
p11.2
B
C
D
EGFRCEP7
EGFR 12 13 14 15 16 Iso A (1210aa)
BT340
EGFR
Supplemental Figure 3
Chr. 7 Chr. 7
p12.1 p11.1p11.2
Two distinct deletions spanning ex. 14-15

1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 161718
1920
2122
X
T/N
cop
y ra
tio
0.5
1
2
Subclonal
Homozygous deletion
Hemizygous deletion
Trisomic
Tetrasomic
Disomic
Clonal54
1
2
0.5
1.5
0
T/N
cop
y ra
tio
a
a. Subclonal deletions in chrs. 1, 4, 6
EGFR
CDKN2A/2B
BA Bulk copy number of BT325 Clonality analysis of SCNA in BT325
Supplementary Figure 4

AGermline heterozygous site
Reference baseAlternate base
Genotype
Possibe haplotypes
~1Kb
Cluster 1 Cluster 2
Clustering based on pairwise distance
Haplotype inference
Retained allelotypeDeleted allelotype
LOH populationHeterozygous population
RD
R R D R D
. . R R . . R
. . . . .. . . . .
1
2
3
4
5
6
Single-cell allelotyping
D D RD D R
. . . R . . R
R R . . R ..
D
0
0.5
1
Allelotype ratio (deleted vs. retained) within SNS
Allelotype distribution within two clusters
Cluster 1
Cluster 2
RD
RD
T/N
mix
dipl
oid
F Cell
B
A
Loss -of-heterozygosity
Haplotypes
Heterozygous
Genotypes from incompletesingle-nucleus sequencing
Homozygous loss
A
B
Genotype from completesingle-nucleus sequencing
1
2
3
4
5
6
Pairwise distance
2/3
C
cell 3 (LOH) vs.
cell 1(Het)
cell 3 (LOH) vs.
cell 4 (LOH)0
Pairwise comparison
E
G
0
0.01
0.02
0.03
0.04
0.05% of bases containing at least 1 error read avg: 0.019; std. dev: 0.005 % of reads containing at least 1 error base avg: 0.012; std. dev: 0.003
Tumor cells with chr. 10 deletion
H
Supplemental Figure 5

1
0.5
0
Haplotype ratio in Chr. 10 (Deleted vs. Retained)
Chromosome 7 copy ratio
Cells with less even coverageCells with more even coverage
0
1
2
3
Normal (diploid)
Tumor cells with complete 6q deletionTumor cells retaining multiple 6q segments
A
B
C
Chromosome 10 deletion
Chromosome 7 amplification
Chromosome 6 haplotypingChr. 9
haplotyping
D E
Haplotype compositions in regions with subclonal deletions
Retained haplotype Deleted haplotype
0 0.5 1 0 1000
Haplotype ratio
D:RTotal # of SNPs
53.6--57 Mb 63.3--66.1 Mb
27.1-30.9 Mb31.8-32.0 Mb
28--33 Mb
Clonal Events
Subclonal deletions in 6q and 9p Subclonal Chr. 16 loss
Subclonal Events
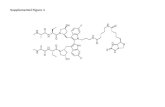
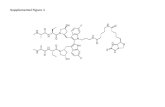
Supplemental Figure 6
0 1
Haplotype ratioD:R
Chr
. 16
LOH
Chr
. 16
non-
LOH
CDKN2A hom. lossChr.10 hem. loss
Deletions in Chrs. 6 and 9
Chr. 16 loss

A
Chr9: 9--35Mb
Retained allelotype Deleted allelotype
B
0 0.5 1
Haplotype ratio (Deleted vs. Retained)
0 0.5 1 1.5 2
Chr. 7 copy ratio
C9p deletion Chr. 10 deletion Trisomy 7
Haplotyping
Clonal Events
Chr7: 54560159-54560356 Chr7: 55510459-55510566
Tumor/normal mixture 1
Tumor/normal mixture 2
E
Amplicon 7: 54,560,250--55,510,789
F
Tumor/normal mixture 1
Tumor/normal mixture 2
Chr3: 22615037-22615438Chr9: 21029084-21029485
Presence of EGFR amplification in T/N mixtures
Presence of Chr. 9-Chr. 3 translocation in T/N mixtures
D
1B1A
8B8A
9B9A
Rearranged segments by the reference assembly
Rearranged segments in derivative chromosomes
Complex rearrangement disrupting CDKN2A
Bulk210
CN
RepresentativeSNS library
Pool ofSNS libraries
Chr. 1
Chr. 8
Chr. 9
Chr. 3 fragments
20,900--22,150Kb
CDKN2A/CDKN2B
9A 8B
1A 9B
8A 1B
der (1,9)
der (8,1)
der (9,8) × 2
Supplementary Figure 7

Chr. 2
Chr. 8
2:187389362
2:187389633
8:32908298
8:32909007
2:1892626238:2261740
2:1894072648:2261928A
8p-tel.
2q-tel.
8p-tel.
8:32908298
B-a5 • -a6 • -a1
a7 • -a3 • -a4 • a9 • a8 • -a2 • -a10
Chr. 12
a b c de f g12a • 12e • 12b • 12f • -12d • 12c • 12f
C Chr. 14
a
b
c d
e
f g nlkjih
m14a • -14l • 14g • -14f • 14d • 14c • 12f
? • 14i • -14j • 14k • 14h • ?
D
Chr. 9Chr. 6 9B6A
Simple deletion
Chr.6 Chr.9 CEP6
Chr. 9
20 25 30 35 40
E
Bulk CN50 60 7055 65
FISH probes Chr. 6Bulk CN
Chr.6 Chr.9 CEP9
extrachromosomal
amplicons
FISH probes Chr. 9chr6:71,726,042-71,726,935 chr9:27,138,441-27,140,176
+
connects to the Chr. 6 amplicon
connects to the Chr. 6 segment
connects to another Chr. 6 segment
connects to another Chr. 6 amplicon
Non-amplified Chr. 6 segment Non-amplified Chr. 9 segment
Chr. 6 amplicon Chr. 9 amplicon
Inferred derivative chromosome in cluster 1
9B6A
9B
Inferred derivative chromosome in cluster 2
6A 9B
6A
Chromothripsis
6.5Mb
9c
9b
6d
6c
6b6a
dm 6:9
Supplemental Figure 8

Chromosome 1 (Mb) Chromosome 4 (Mb) Chromosome 6 (Mb)
20 22 24 26 183 185 187 110 120 130 140 150 160
0.5
Min
or a
llele
fre
quen
cy
0
1
Bulk CN
0.50 1 1.5
All tumor nuclei
Haplotype ratio (Deleted vs. Retained)
Nuclei retaining heterozygosity
Nuclei having lostheterozygosity
Two nucleus mixture
A
Retained allelotype Deleted allelotype
0.5
01
1.5
Nuclei retaining heterozygosity
Nuclei having lostheterozygosity in chr. 1, 4, 6
1:1 mixture
00.
51
Chr. 10
Deletions in chrs 1, 4, 6
Tumor nuclei Non-tumor nuclei
CDKN2A homozygous deletionChr. 10 hemizygous deletion Heterozygous Chr. 10
Diploid normal 1:1 mixture T/N
Hap
loty
pe r
atio
D:R
Hap
loty
pe r
atio
D:R
Chromosome 1 (Mb) Chromosome 4 (Mb) Chromosome 6 (Mb)20 22 24 26 183 185 187 110 120 130 140 150 160
0.5
Min
or a
llele
fre
quen
cy
0
1
Bulk CN
LOH tumor nuclei only
1a 1b
1A4a 4b
4A4A4A
6a 6b 6c 6d 6e 6f 6h6g 6i
6D 6E 6F6G
Rearranged segments by the reference assembly
Rearranged segments in derivative chromosomes
E
Chr. 1
Chr. 4
Chr. 6
4a 4b
1a
6a
1b6i
6e6b
6c6f
6h
6g
der 1:6
der 4
der 6:1
6d
Supplementary Figure 9
F
D
B C

0
50
100
150
0
50
100
150
100
50
150
0
0
50
100
150
Per
cent
Sur
viva
lP
erce
nt S
urvi
val
Per
cent
Sur
viva
lP
erce
nt S
urvi
val
C D
Erlotinib (nM)10 10 2 10 3 10 4
10 10 210 3 10 41
Lapatinib (nM)
0.01 0.1 101 101 100Neratinib (nM)Afatinib (nM)
vII
vIII
Vector + IL-3
WT + IL-3
EGFR vII EGFR vIII
IC50(nM) 4.37.8
vII
vIII
Vector + IL-3
WT + IL-3
EGFR vII EGFR vIII
IC50(nM) 930210EGFR vII EGFR vIII
IC50(nM) 8455
EGFR vII EGFR vIII
IC50(nM) 0.480.62
A B
Supplemental Figure 10: