Supplementary Figure 1. Examination of neural cells and Wnt … · 2012-08-17 · Supplementary...
Transcript of Supplementary Figure 1. Examination of neural cells and Wnt … · 2012-08-17 · Supplementary...

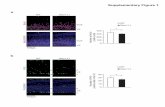
Supplementary Figure 1. Examination of neural cells and Wnt signaling in differentiating
H9 hESC cultures at day 4 of UM treatment.
(a) Flow cytometry distribution of differentiating H9 hESCs at day 4 of UM treatment. Green dots
indicate nestin+/βIII tubulin- events, red dots indicate nestin+/βIII tubulin+ events, blue dots
indicate nestin-/βIII tubulin+ events, and black dots indicate nestin-/βIII tubulin- events. (b)
Expression of WNT7A and WNT7B detected by RT-PCR at 4 days of UM treatment. (c-d)
Expression of nestin (c) and WNT7A (d) were detected by combined fluorescence in situ
Nature Biotechnology: doi:10.1038/nbt.2247

hybridization/immunocytochemistry. (e-f) Expression of βIII tubulin (e) and WNT7A (f). (g-h)
Expression of nestin (g) and WNT7B (h). (i-j) Expression of βIII tubulin (i) and WNT7B (j). Scale
bars indicate 50 µm. Adjacent horizontal panels represent the same field.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 2. Identification of GFAP+ and α-SMA+ cells in the differentiating
IMR90-4 cultures.
(a) Glial fibrillary acidic protein-positive (GFAP+) cells (red) could be identified among the βIII
tubulin+ cells (green) by immunocytochemistry at day 6 of UM treatment. (b) α-smooth muscle
actin (α-SMA+) cells (green) were also identified and are shown overlaid with DAPI nuclear stain
(blue). Scale bars indicate 50 µm. Neither cell type was present in significant quantities.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 3. IMR90-4-derived BMECs express requisite BBB markers after
continued differentiation in EC medium.
IMR90-4-derived BMECs were cultured for 6 days in UM and 2 days in EC medium prior to
immunolabeling. (a-b) PECAM-1 (a) co-expressed with claudin-5 (b). (c) Expression of occludin.
(d) Expression of GLUT-1. (e) Expression of p-glycoprotein. All scale bars indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 4. DF19-9-11T-derived BMECs express requisite BBB markers.
DF19-9-11T-derived BMECs were cultured for 6 days in UM and 2 days in EC medium prior to
immunolabeling. (a-b) PECAM-1 (a) is shown co-expressed with claudin-5 (b). (c) Expression
of GLUT-1. (d) Expression of p-glycoprotein. All scale bars indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 5. DF6-9-9T-derived BMECs express requisite BBB markers.
DF6-9-9T-derived BMECs were cultured for 6 days in UM and 2 days in EC medium prior to
immunolabeling. (a-b) PECAM-1 (a) is shown co-expressed with occludin (b). (c-d) GLUT-1 (c)
is shown co-expressed with claudin-5 (d). (e) Expression of p-glycoprotein. All scale bars
indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 6. H9-derived BMECs express requisite BBB markers.
H9-derived BMECs were cultured for 7 days in UM and 6 days in EC medium prior to
immunolabeling. (a-b) PECAM-1 (a) is shown co-expressed with claudin-5 (b). (c) Expression of
occludin. (d) Expression of GLUT-1. (e) Expression of p-glycoprotein. All scale bars indicate 50
µm.
Nature Biotechnology: doi:10.1038/nbt.2247

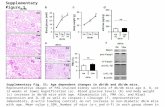
Supplementary Figure 7. Examination of neural cells and Wnt signaling in differentiating
IMR90-4 iPSC cultures.
(a) Expression of WNT7A and WNT7B detected by RT-PCR at 4 days of UM treatment. (b-c)
Expression of nestin (b) and WNT7A (c) were detected by combined fluorescence in situ
hybridization/immunocytochemistry at day 6 of UM treatment. (d-e) Expression of βIII tubulin (d)
and WNT7A (e) were detected at day 6 of UM treatment. Arrowheads indicate a cell with
positive expression of βIII tubulin that lacks WNT7A expression. Panels (b) and (c), and panels
(d) and (e), are the same field. Scale bars indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 8. DF19-9-11T, DF6-9-9T, and H9 cell lines demonstrate β-catenin
nuclear localization during BMEC specification.
(a) After 6 days in UM and 2 days in EC medium, DF19-9-11T-derived BMECs were
immunolabeled for PECAM-1 expression (red) and β-catenin (green). (b) After 6 days in UM and
2 days in EC medium, DF6-9-9T-derived BMECs were immunolabeled for GLUT-1 (red) and β-
catenin (green). (c) After 7 days in UM and 3 days in EC medium, H9-derived BMECs were
immunolabeled for GLUT-1 (red) and β-catenin (green). H9-derived ECs required a longer time
frame to acquire nuclear β-catenin, which may explain the mixture of BBB and non-BBB ECs.
Scale bars represent 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 9. DF19-9-11-derived BMECs subcultured onto
collagen/fibronectin-coated surface maintain BBB markers.
(a) vWF (red) is shown co-localized with occludin (green) by immunofluorescence. DAPI (blue)
is overlaid. (b) Expression of PECAM-1 (red) overlaid with DAPI (blue). (c) Expression of
claudin-5. (d) Expression of GLUT-1. (e) Expression of p-glycoprotein. All scale bars indicate 50
µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 10. DF6-9-9-derived BMECs subcultured onto collagen/fibronectin-
coated surfaces maintain BBB markers.
(a-b) PECAM-1 (a) is shown co-expressed with claudin-5 by immunofluorescence (b). DAPI
(blue) is shown overlaid with PECAM-1. (c) Expression of occludin. (d) Expression of GLUT-1.
(e) Expression of p-glycoprotein. All scale bars indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 11. Continuous tight junction expression is maintained by IMR90-
4-derived BMECs after subculture onto collagen/fibronectin-coated Transwell® filters.
IMR90-4-derived BMECs were differentiated for 6 days in UM and 2 days in EC medium prior to
subculture onto collagen/fibronectin-coated filters. Immunofluorescence images were recorded
once cells reached confluence after 2 days. (a) claudin-5. (b) occludin. (c) ZO-1. All scale bars
indicate 50 µm.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 12. Stability of the IMR90-4-derived BMEC phenotype over
extended culture times
IMR90-4-derived BMECs were co-cultured with rat astrocytes and TEER was measured
approximately every 24 hours. TEER values are normalized to the maximum value within each
experiment. The 1% PDS time course is the same as that documented in Figure 4b, and the
10% FBS time courses are demonstrating the consistent maximum and plateau time course
profiles for three independent biological replicates.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Figure 13. Basolateral to apical transport of rhodamine 123 decreases in
the presence of cyclosporin A.
Basolateral to apical transport of rhodamine 123 decreases in the presence of cyclosporin A
(80±5%) compared to the control (100±4%). Statistical significance was calculated by the
Student’s t-test; *, p<0.07.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Table 1. In vitro permeability of the IMR90-4-derived BMECs.
Compound Molecular Weight
(g/mol) Octanol/water partition
coefficient (logD)1-6 Transporter
recognition7-10 Average Pe
(10-3 cm/min)b
Inulin Variablea -0.25 None 0.029 ± 0.02
Sucrose 342 -3.3 None 0.034 ± 0.015
Glucose 180 -2.8 GLUT-1 (influx) 0.22 ± 0.07
Vincristine 825 2.8 MDR1, MRP1,
MRP2 0.062 ± 0.02
Colchicine 399 1.03 MDR1, MRP1 0.092 ± 0.1
Prazosin 383 2.16 BCRP (efflux), Organic cation
transporters (influx) 0.29 ± 0.04
Diazepam 285 2.82 None 1.1 ± 0.5 aInulin is a polysaccharide polymer with an unspecified chain length. bMean ± standard deviation was calculated from three completely independent biological replicates except sucrose (eighteen replicates).
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Table 2. Efflux transporter substrates and inhibitors.
Compounds11-13 Transporter recognition11-13
Substrates
Rhodamine 123 MDR1
Doxorubicin MDR1, BCRP, MRP1, MRP2
Inhibitors
Cyclosporin A MDR1
Ko143 BCRP
MK 571 MRP family
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Table 3. Antibodies used for immunofluorescence and flow cytometry.
Targeted antigen Antibody description Vendor Dilution
PECAM-1 Polyclonal rabbit Thermo Fisher 1:25
von Willebrand Factor Polyclonal rabbit Dako 1:100
VE-cadherin Mouse monoclonal SCBT 1:100
ZO-1 Polyclonal rabbit Invitrogen 1:100
Occludin Mouse monoclonal Invitrogen 1:50
Claudin-5 Mouse monoclonal Invitrogen 1:200
GLUT-1a Polyclonal rabbit antiserum N/A 1:500
GLUT-1a Mouse monoclonal Thermo Fisher 1:100
p-glycoprotein Mouse monoclonal Thermo Fisher 1:25
β-catenin FITC-conjugated mouse
monoclonal BD Biosciences 1:150
βIII tubulin Polyclonal rabbit Sigma 1:500
Nestinb Polyclonal rabbit Millipore 1:1000
Nestinb Mouse monoclonal Millipore 1:500
GFAP Polyclonal rabbit Dako 1:500
α-smooth muscle actin Mouse monoclonal ARP 1:100
Digoxigenin Mouse monoclonal Sigma 1:500
K14 Rabbit polyclonal Thermo Fisher 1:100
K18 Mouse monoclonal Thermo Fisher 1:100
p63 Mouse monoclonal SCBT 1:33 aThe polyclonal GLUT-1 antiserum7 was used for immunofluorescence only. The monoclonal GLUT-1 antibody was used for immunofluorescence and flow cytometry. bThe polyclonal nestin antibody was used for immunofluorescence and the monoclonal nestin antibody was used for immunofluorescence and flow cytometry.
Nature Biotechnology: doi:10.1038/nbt.2247

Supplementary Table 4. Primers used for RT-PCR and qPCR. Genea Forward sequence Reverse sequence GAPDH CACCGTCAAGGCTGAGAACG GCCCCACTTGATTTTGGAGG SLC2A1 ACGCTCTGATCCCTCTCAGT GCAGTACACACCGATGATGAAG ABCB1 TGAATCTGGAGGAAGACATGAC CCAGGCACCAAAATGAAACC LEF1 CAGATGTCAACTCCAAACAAGG GATGGGATATACAGGCTGACC
STRA6 TTTGGAATCGTGCTCTCCG AAGGTGAGTAAGCAGGACAAG FZD4 TACCTCACAAAACCCCCATCC GGCTGTATAAGCCAGCATCAT FZD6 TCGTCAGTACCATATCCCATG CCCATTCTGTGCATGTCTTTT FZD7 GATGATAACGGCGATGTGA AACAAAGCAGCCACCGCAGAC FST GTTCATGGAGGACCGCAGTG TCTTCTTGTTCATTCGGCATT
APCDD1 GGAGTCACAGTGCCATCACAT CCTGACCTTACTTCACAGCCT PECAM1 GAGTATTACTGCACAGCCTTCA AACCACTGCAATAAGTCCTTTC
CDH5 CGCAATAGACAAGGACATAACAC GGTCAAACTGCCCATACTTG VWF CCCGAAAGGCCAGGTGTA AGCAAGCTTCCGGGGACT LDLR GCCATTGTCGTCTTTATGTC AAACACATACCCATCAACGA
SLC7A5 TTAAAGTAGATCACCTCCTCGA GGATGAGATTCGTACCAGAG SLC16A1 GGTGTTTCTTAGTAGTTATGGG TCTTATTGGCTTTGTGTTGG
INSR TGTTCATCCTCTGATTCTCTG GCTTAGATGTTCCCAAAGTC LEPR GGAAATCACACGAAATTCAC GCACGATATTTACTTTGCTC BCAM GCTTTCCTTACCTCTAAACAG GAAGGTGATAGAACTGAGCG
SLC38A5 TGTCAGTGTTCAACCTCAG GTGGATGGAGTAGGACGA SLC1A1 GTTATTCTAGGTATTGTGCTGG CTGATGAGATCTAACATGGC ABCG2 TCAGGTCTGTTGGTCAATCTC GTTTCCTGTTGCATTGAGTCC ABCC1 AATAGAAGTGTTGGGCTGAG CGAGACACCTTAAAGAACAG ABCC2 ATATAAGAAGGCATTGACCC ATCTGTAGAACACTTGACCA ABCC4 AATCTACAACTCGGAGTCCA CAAGCCTCTGAATGTAAATCC ABCC5 TCACTACATTAAGACTCTGTCC GGATACTTTCTTTAGGACGAGAG AGER GTAGATTCTGCCTCTGAACTC CTTCACAGATACTCCCTTCTC
SLC21A14b AAAGATGTGGAAGTAGAGGA ATGCTTAGGAGAATTGACAC PLVAPb CAATGCAGAGATCAATTCAAGG ACGCTTTCCTTATCCTTAGTG TFRC GCACAGCTCTCCTATTGAAAC GGTATCCCTCTAGCCATTCAG LRP1 GACTACATTGAATTTGCCAGCC TCTTGTGGGCTCGGTTAATG
WNT7A CGGGAGATCAAGCAGAATG CGTGGCACTTACATTCCAG WNT7B GCTTCGTCAAGTGCAACA GGAGTGGATGTGCAAAATG WNT7Ac TGGAACAGAATAGTTGAGGGCT N/A WNT7Bc AGCCAAGGGACAGTGCGAGTGT N/A
aFor genes with multiple transcript variants (e.g. LEPR, BCAM), the primers were designed to encompass all transcript variants and not one single transcript isoform. bFidelity of SLC21A14 and PLVAP primers was confirmed using HUVECs because these genes were not expressed in the IMR90-4-derived BMECs. cDIG-labeled locked nucleic acid probe sequences used for fluorescence in situ hybridization. DIG-labeled locked nucleic acid sense probes were used as controls.
Nature Biotechnology: doi:10.1038/nbt.2247

References 1. Huang, J.D. Comparative drug exsorption in the perfused rat intestine. The Journal of
pharmacy and pharmacology 42, 167-170 (1990). 2. Mazzobre, M.F., Roman, M.V., Mourelle, A.F. & Corti, H.R. Octanol-water partition
coefficient of glucose, sucrose, and trehalose. Carbohydrate research 340, 1207-1211 (2005).
3. Ghasemi, J. & Saaidpour, S. Quantitative structure-property relationship study of n-octanol-water partition coefficients of some of diverse drugs using multiple linear regression. Analytica chimica acta 604, 99-106 (2007).
4. Levin, V.A. Relationship of octanol/water partition coefficient and molecular weight to rat brain capillary permeability. Journal of medicinal chemistry 23, 682-684 (1980).
5. Quinn, F.R. & Beisler, J.A. Quantitative structure-activity relationships of colchicines against P388 leukemia in mice. Journal of medicinal chemistry 24, 251-256 (1981).
6. Jones, D.R., Hall, S.D., Jackson, E.K., Branch, R.A. & Wilkinson, G.R. Brain uptake of benzodiazepines: effects of lipophilicity and plasma protein binding. The Journal of pharmacology and experimental therapeutics 245, 816-822 (1988).
7. Pardridge, W.M., Boado, R.J. & Farrell, C.R. Brain-type glucose transporter (GLUT-1) is selectively localized to the blood-brain barrier. Studies with quantitative western blotting and in situ hybridization. The Journal of biological chemistry 265, 18035-18040 (1990).
8. Szakacs, G., Paterson, J.K., Ludwig, J.A., Booth-Genthe, C. & Gottesman, M.M. Targeting multidrug resistance in cancer. Nat Rev Drug Discov 5, 219-234 (2006).
9. Hayer-Zillgen, M., Bruss, M. & Bonisch, H. Expression and pharmacological profile of the human organic cation transporters hOCT1, hOCT2 and hOCT3. British journal of pharmacology 136, 829-836 (2002).
10. Cisternino, S., Mercier, C., Bourasset, F., Roux, F. & Scherrmann, J.M. Expression, up-regulation, and transport activity of the multidrug-resistance protein Abcg2 at the mouse blood-brain barrier. Cancer research 64, 3296-3301 (2004).
11. Morjani, H. & Madoulet, C. Immunosuppressors as multidrug resistance reversal agents. Methods in molecular biology (Clifton, N.J 596, 433-446.
12. Perriere, N. et al. A functional in vitro model of rat blood-brain barrier for molecular analysis of efflux transporters. Brain Res 1150, 1-13 (2007).
13. Wang, Q., Yang, H., Miller, D.W. & Elmquist, W.F. Effect of the p-glycoprotein inhibitor, cyclosporin A, on the distribution of rhodamine-123 to the brain: an in vivo microdialysis study in freely moving rats. Biochemical and biophysical research communications 211, 719-726 (1995).
Nature Biotechnology: doi:10.1038/nbt.2247