Supplemental Information: Endothelial Hypoxia Inducible Factor-2alpha (HIF...
Transcript of Supplemental Information: Endothelial Hypoxia Inducible Factor-2alpha (HIF...

1
Supplemental Information:
Endothelial Hypoxia Inducible Factor-2alpha (HIF-2α) regulates murine
pathological angiogenesis and revascularization processes
Nicolas Skuli1,2, Amar J. Majmundar2,3, Bryan L. Krock2, Rickson C. Mesquita4, Lijoy K. Mathew1,2,
Zachary L. Quinn1,2, Anja Runge1,2,5, Liping Liu1,2,6, Meeri N. Kim4, Jiaming Liang4, Steven
Schenkel4, Arjun G. Yodh4, Brian Keith1,7, M. Celeste Simon1,2,8
1Howard Hughes Medical Institute, 2Abramson Family Cancer Research Institute, 3School of
Medicine, 4Department of Physics & Astronomy, 7Department of Cancer Biology, 8Department of
Cell and Developmental Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Present Affiliation and location: 5Division of Vascular Oncology and Metastasis, German Cancer Research Center, Heidelberg, D-
69120, Germany. 6Merck Research Laboratories, West Point, PA 19486, USA.
Corresponding author: M. Celeste Simon, PhD Scientific Director and Investigator, Abramson Family Cancer Research Institute Investigator, Howard Hughes Medical Institute Professor, Cell and Developmental Biology University of Pennsylvania School of Medicine 456 BRB II/III, 421 Curie Boulevard Philadelphia, PA 19104-6160 Telephone number: 215-746-5532 Fax number: 215-746-5511 E-mail: [email protected] Conflict of interest: The authors declare no competing financial interests

2
Supplemental Information: Methods
Antibodies
Antibodies were purchased from commercial sources and used according to
manufacturers’ instructions. Specifically: CD31 (Clone MEC13.3), ICAM-2, biotin-CD31, biotin-
ICAM-2, Fc Block (BD Pharmingen) ; Von Willebrand factor (Millipore) ; Actin, Dll4, Ang2, NICD,
Desmin (Abcam) ; Ang2 (Santa Cruz) ; smooth muscle actin (clone 1A4), FITC-smooth muscle
actin (Sigma) ; HIF-1α (Transduction Labs and Novus) and HIF-2α (Novus) ; Ki67 (BD
Biosciences) ; Horseradish peroxidase-labeled goat anti-rabbit IgG or anti-mouse IgG (Cell
signaling). Alexa 488/546 conjugated anti-mouse, rabbit, and goat IgG were purchased from
Invitrogen.
Speckle imaging, blood flow and oxygenation assessment
DCS measurements were performed with a home-built instrument containing two
continuous wave, long coherent 785 nm lasers (CrystaLaser Inc., Reno, NV) and eight avalanche
photodiodes (PerkinElmer, Canada). Two 4-channel autocorrelator boards (Correlator.com,
Bridgewater, NJ) were employed to compute the intensity temporal correlation function for an
integration time of 500 ms. Data were collected simultaneously in both limbs, with four detectors
distributed symmetrically along one single source positioned at the center, thus providing two
source-detector separations (0.5 and 1.0 cm) from both the top and bottom of the source position.
Measurements of DCS were also compared with blood flow measurements with a 1-channel LDF
(BPM 403A, TSI Inc., St. Paul, MN). In order to compare flow from the same region over the two
different techniques, we measured 3 different points along the bottom portion of the DCS probe,
symmetrically positioned in each limb.
The DRS system was composed of a 250 W tungsten-halogen light source (TS-428, PI
Acton, MA), a spectrometer (SP-150, Acton Research, MA), and a 16-bit, back-illuminated CCD
camera (PIXIS:400BR, Princeton Instruments, NJ). The optical probe consisted of a single source
and ten detection fibers spaced non-uniformly (0.6, 1.2, 1.8, 2.4, 3, 4, 5, 6, 8, and 10 mm). A

3
nonlinear fitting procedure was employed to simultaneously fit all data in the optimal wavelength
range (600 – 850 nm) using the analytical solution of the photon diffusion equation for a semi-
infinite medium with extrapolated zero boundary conditions. For each limb measurement, we
collected data at 3 different locations along the limb, and 10 different frames at every location,
with an exposure time of 100 ms each.
All DCS, LDF and DRS measurements were performed prior to left hindlimb artery
occlusion, in random order. The same set of measurements was repeated right after surgery, and
subsequently once a week, up to maximum of four weeks, when the animals were sacrificed.
Quantitative real-time PCR analysis
Applied Biosystem Primer references:
Dll4: Mm00444619_m1 (RefSeq: NM_019454.3)
Hey1: Mm00468865_m1 (RefSeq: NM_010423.2)
Hey2: Mm00469280_m1 (RefSeq: NM_013904.1)
Hes1: Mm00468601_m1 (RefSeq: NM_008235.2)
Hes2: Mm00456108_g1 (RefSeq: NM_008236.4)
Notch1: Mm00435245_m1 (RefSeq: NM_008714.3)
ADM1: Mm00437438_g1 (RefSeq: NM_009627.1)
Tie2: Mm00443254_m1 (RefSeq: NM_013690.2)
Ang1: Mm00456503_m1 (RefSeq: NM_009640.3)
Ang2: Mm00545822_m1 (RefSeq: NM_007426.3)
EphrinB2: Mm01215897_m1 (RefSeq: NM_010111.5)
EphrinB4: Mm01201157_m1 (RefSeq: NM_001159571.1)
VEGF: Mm01281449_m1 (RefSeq: NM_001025250.3)
Immunoblotting
Endothelial cells were incubated under normoxia (21% O2) or hypoxia (0.5% O2) for 16
hrs. Cells were harvested and lysed in RIPA buffer. 40 to 80 µg of proteins was loaded on 8%
acrylamide gels. After completing SDS-PAGE and electrophoretic transfer for 4 hrs at 4°C,

4
membranes were blocked for 1 hr in 5% Non Fat Dry Milk (BD Pharmingen) in TBS with 0.05%
tween-20 (Sigma). Blots were probed overnight at 4°C with the appropriate antibody and
following the manufacturers’ instructions. Horseradish peroxidase-labeled goat anti-rabbit IgG or
anti-mouse IgG (Cell Signaling) were used, respectively, at 1:10,000 and 1:5,000.
Flow cytometry
Control and KO endothelial cells were prepared as a single cell suspension and washed
in FACS buffer (PBS, 5% FCS, 0.5% BSA). Cells were blocked for 10 min using a blocking
antibody (Fc Block, BD Pharmingen) in FACS buffer and stained using primary antibodies (CD31,
ICAM2 and VWF) for 45 min at 4°C. After washes, cells were incubated with the appropriate
secondary antibodies for 30 min at 4°C. Finally, cells were washed twice, resuspended in 500 µl
of isotone (Coulter Electronics) and analyzed using BD FACS Calibur system, Flowjo software
(Tree Star Inc) and controls with isotype-matched IgG.
Endothelial cells with HIF-1α /HIF-2α acute deletion (Cnt.H1.2/KO.H1.2 and
Cnt.H2.2/KO.H2.2)
Lungs from Hif-2αfl/Δ / Ubc-CreER and Hif-1αfl/fl / Ubc-CreER mice were isolated and
processed as described above to obtain endothelial cells. This Cre-ER transgene is expressed
from the human Ubiquitin C promoter encoding the Cre protein fused to a modified form of the
estrogen receptor that specifically binds Tamoxifen with high affinity and is inactive until
Tamoxifen is administered to the cells (Rusankina et al., 2007, Cell Stem Cell 1:113-26). Cre-ER
recombinase was activated in cells by adding Tamoxifen (1µM, MP Biomedicals) in culture
media. DNA extracted from cells shows that recombination is highly efficient.
Phalloidin Staining
Control and KO cells were rinsed in PBS briefly to remove media components and fixed
in 1% paraformaldehyde in PBS (freshly prepared) for 15 min at RT. Aldehyde excess was
quenched with 10 mM ethanolamine in PBS for 5 min. Cells were permeabilized in 0.1% Triton-

5
X100 in PBS for 1 min and incubated in FITC-phalloidin (Molecular Probes) diluted 1:100 in PBS
for 15 min. Finally, cells were rinsed 3 times in PBS, 5 min/wash and mounted for microscopy.
Cell proliferation assay
Control and KO cells were seeded into 6 cm dishes at 5x104 cells per well and incubated
at 21% O2 or 0.5% O2 for 5 days. Every day, cells were harvested, washed in PBS and counted
using a hemocytometer.
BrdU and cell death assay
Control and KO cells were pulsed with BrdU (10µM) for 30 minutes. The cells were
processed for FACS analysis according to manufacturer’s instructions (BD Biosciences). To
analyze the percent of cells undergoing cell death, FACS analysis was performed using Annexin
V-FITC and PI staining (BD Biosciences).
Invasion assay
Cells were plated in serum-free EGM media (Lonza) on 24-well Transwell inserts (Costar,
Corning). The lower chamber contained complete EGM media; after incubation for 8 hrs at
37 °C/5% CO2/21% O2 or 0.5% O2, the inserts were fixed with 3.7%
paraformaldehyde/phosphate-buffered saline and stained with 2% crystal violet. The number of
invaded cells per 100X field was determined using a Leica 500 microscope (Leica).
Viability assay
We assessed the percentage of viable Cnt. and KO cells in different culture conditions
(+/- O2 and +/- serum). A 1x106 cells/ml cell suspension was prepared and diluted using 1:1 ratio
of 0.4% trypan blue solution (Cellgro). Stained cells and total number of cells were determined by
hemocytometer counting. The calculated percentage of unstained cells represent the percentage
of viable cells.

6
Collagen adhesion assay
Adhesion assays were performed as described (52). Briefly, KO and related Cnt. ECs
were seeded into 24-well plates coated or not with Collagen (1 µg/ml, Upstate) and incubated for
1hr at 37°C under normoxic or hypoxic conditions. Cells were then carefully washed 3 times with
PBS, fixed and counted. The number of adherent cells was counted in 6 representative HPFs
(high power fields) using an Olympus IX81 microscope (Olympus). The data are representative of
three independent experiments.

Ctrl KO
0.3
0.2
Rat
ioPe
ricy
te/E
C
0
Ligated limb
0.1
Ligated
KO
Ctrl
A DAPI Desmin CD31 Merged
Skuli et al., Supplemental Figure 1
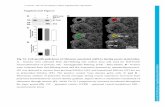
Supplemental Figure 1. Defective smooth muscle cell coverage in mice with endothelial specific HIF-2αα deletion. (A) Imagesshowing desmin (pericyte marker) and CD31 (endothelial cell marker) co-staining in ligated limb from Ctrl and KO mice. Scale bars: 40 µm.Magnification 200X. (B) Pericyte recruitment is expressed as the ratio between pericyte to EC and no significant (N.S.) difference isobserved between Ctrl and KO mice. (Ctrl n=8, KO n=9). (C) Pictures showing SMC staining in adductor muscle for ligated and nonligated limb in Ctrl and KO mice. Scale bars: 40 µm. Magnification 200X. (D) Quantification of number of α-SMA-stained vessels perfield showing no significant difference between Ctrl and KO mice. (E-F) In correlation with collateral artery thickness wall reduction,defective smooth muscle cell coverage was observed in KO mice. Representative pictures (E) showing SMC staining in adductor mus-cle for ligated and non ligated limb in Ctrl and KO mice. SMC coverage per vessel area was assessed (F). Scale bars: 20 µm. Magnifi-cation 400X. (Ctrl n=8, KO n=9). (G-H) Images showing SMC staining (G) in skin tumors from Ctrl and KO mice. SMC coverage per vesselarea was assessed (H). Scale bars: 20 µm. Magnification 400X. (Ctrl n=10, KO n=11). Data are means ± SEM. *P<0.05.
Ctrl KO0
100
SM
Ccovera
ge
(µm
2 )
150
50
*
G KOCtrl
Ctrl KO
10
25
20
15
No.of
α-SM
A-s
tain
edve
ssel
s/fiel
d
0
Ligated limbNon ligated limb
5
D
KO
Ctrl
Non-ligated LigatedC
Ctrl KO
250
400
350
300
0
Ligated limbNon ligated limb
100
SM
Cco
vera
ge
(µm
2 )
150
200
50
*
KO
Ctrl
Non-ligated LigatedE
F H
α−SMA α−SMA α−SMA
N.S.
B

CtrlH2.2
KOH2.2
+Tam1µM
fl
∆wt
CtrlH2.1
KOH2.1
C
Ctrl H2.1 KO H2.1
Unstained
ICAM2
VWF
CD31
B
GFP GFP
LDL uptake
Ctrl H2.1 KO H2.1MS 1ASkuli et al., Supplemental Figure 2
HIF-2α
HIF-1α
N HCtrl H2.1 KO H2.1
N H
Actin
N HCtrl H2.2 KO H2.2
N H
+Tam 1µME
120 kD
110 kD
N HCtrl H1.1 KO H1.1
N H N HCtrl H1.2 KO H1.2
N H
+Tam 1µMF
D
MS1 Ctrl H1.1KO H1.1
fl∆
wt
Ctrl H1.2 KO H1.2
+Tam1µM
Supplemental Figure 2. Characterization of HIF-1αα and HIF-2αα KO endothelial cells. (A) Fluorescent immunostaining foracetylated-LDL uptake was studied using MS1, a murine pancreatic endothelial cell line, Ctrl and KO lung endothelial cells. (B) Fluores-cent immunostaining (right panels) and flow cytometric analyses (left panels) were perfomed for CD31, ICAM2 and von Willebrand Fac-tor using Ctrl and KO lung endothelial cells. Scale bars: 5 µm. Magnification 400X. (C-D) PCR reactions showing the efficient recombi-nation in the HIF-1α and HIF-2α KO endothelial cells. (E-F) Western blot analysis of HIF-1α and HIF-2α protein expression in Ctrl andKO endothelial cells subjected to 21% (N) or 0.5% (H) O2 for 6 hrs. The bands appear diffuse due to phosphorylation and other post-translational modifications. Experiments were performed in triplicates.
HIF-2α
HIF-1α
Actin
MS1

fl
∆wt
Muscle ECs Skin ECs
Ctrlfl/wt
KOfl/∆
Ctrlfl/wt
KOfl/∆
A
Skuli et al., Supplemental Figure 3
Unstained ICAM2CD31
MS1
Muscle ECs
Skin ECsCtrlfl/wt
Ctrlfl/wt
Muscle ECsKOfl/∆
KOfl/∆
Skin ECs
B
Supplemental Figure 3. Characterization of skin and muscle HIF-2αα KO endothelial cells. (A) PCR reactions showing the efficientrecombination of the floxed allele in skin and muscle HIF-2α KO endothelial cells. Endothelial cells were isolated from Ctrl (VE-Cad Cre; HIF-2α fl/wt) and KO (VE-Cad Cre ; HIF-2α fl/∆) mice. (B) Flow cytometric analyses were perfomed for CD31 and ICAM2 using Ctrland KO skin and muscle endothelial cells.

E
0.5% O2 Serum-free media
24hrs 48hrs 72hrs 96hrs
20
80
60
40
100
0
Via
bili
ty(%
)
Ctrl H2.1
Ctrl H2.2KO H2.1
KO H2.2
Ctrl H2.1
Ctrl H2.2KO H2.1
KO H2.2
Via
bili
ty(%
)
0.5% O2 Complete media
24hrs 48hrs 72hrs 96hrs
20
80
60
40
0
100
Cell
num
ber
(.1
06
)
0.5
1
1.5
2
2.5
3
Day1 Day2 Day3 Day4 Day50
Ctrl H2.1
Ctrl H2.2
KO H2.1
KO H2.2
0.5% O23.5
BCel
lnum
ber
(.1
06
)
0.5
1
1.5
2
2.5
3
Day1 Day2 Day3 Day4 Day50
Ctrl H2.1
Ctrl H2.2
KO H2.1
KO H2.2
21% O23.5
A
Skuli et al., Supplemental Figure 4
D
Supplemental Figure 4. HIF-2αα deletion does not affect endothelial cell proliferation or viability. (A-B) Ctrl and KO lung ECs wereincubated under 21%(A) or 0.5%(B) O2 in regular culture media and harvested every 24 hrs until the cultures reached confluence. Theaverage cell number of triplicate plates for each condition was determined by counting by hemocytometer following trypsinization. (C)Ctrl and HIF-2α KO ECs were incubated under 0.5%O2 in serum-free media for 48 hrs. Cell cycle was analyzed using BrdU assay.Graph representing the percentage of Ctrl and HIF-2α KO ECs in S after 48hrs under hypoxic conditions (0.5%O2) in serum-free media. No sig-nificant difference in the percentage of ECs in S phase between Ctrl and KO was observed. (D-E) Ctrl and KO ECs were cultured at 0.5%O2in the presence (D) or absence (E) of serum and viable cells were counted at indicated times. Graphs represent the percentage of via-ble cell obtained from the average of three independent experiments. HIF-2α KO ECs in serum-free media show a non-statistically signifi-cant reduction in viability compared to Ctrl ECs. (F) Ctrl and KO ECs were incubated under 0.5%O2 in serum-free media for 48hrs. Celldeath was analyzed using AnnexinV/PI assay. Graph representing the percentage of cell death for Ctrl and KO ECs. No significant differencein cell death between Ctrl and KO ECs was observed. (G-H) Proliferation of ECs was assessed in vivo at day 7 and day 21 after femoral arteryligation for Ctrl and KO mice using Ki67 and Rhodamine lectin co-staining. (G) Images of Ki67 and Rhodamine lectin co-staining showingincreased EC proliferation at day 7 compared to day 21 in adductor muscle, but no significant difference between Ctrl and KO mice.Arrowheads indicate Ki67 positive ECs. Scale bars:20µm. Magnification400X. (H) Quantification of the percentage of Ki67 positiveECs at day 7 and day 21 after femoral artery ligation for Ctrl and KO mice showing no significant difference in EC proliferation in vivobetween Ctrl and KO mice. (Ctrl n=8, KO n=9). Data are means ± SEM.
0.2
0.4
0.6
0.8
1
0
%of
cells
inS
pha
se
CtrlH2.1
CtrlH2.2
KOH2.1
KOH2.2
CBrdU
N.S.
N.S.
Cel
ldea
th(%
)
CtrlH2.1
CtrlH2.2
KOH2.1
KOH2.2
5
10
15
20
30
0
25
FN.S.
N.S.
Annexin V / PI
%of
Ki6
7+
ECs
2
8
6
4
10
0Day7 Day21
CtrlKO
H N.S.
N.S.
CtrlDay7
KODay7
DAPI Ki67 Lectin MergedG
CtrlDay21
KODay21

15
60
45
30
75
0CtrlH2.1
CtrlH2.2
KOH2.1
KOH2.2
Num
ber
ofce
llsat
tach
ed/H
PF
Plastic 21% O20.5% O2
40
160
120
80
200
0CtrlH2.1
CtrlH2.2
KOH2.2
Num
ber
of
cells
atta
ched
/HPF
21% O20.5% O2
Collagen I
**
Phal
loid
in
KO H2.1Ctrl H2.1
Normoxia
KO H2.2Ctrl H2.2
KO H2.1Ctrl H2.1
Hypoxia
KO H2.2Ctrl H2.2
Phal
loid
in
D
A
Skuli et al., Supplemental Figure 5
B
Supplemental Figure 5. HIF-2αα deletion affects endothelial cell adhesion and filopodia formation. (A-B) Reduced adhesion ofindependently derived KO EC lines to plastic (A) or collagen type I- (B) coated plates, under normoxic (21% O2) or hypoxic (0.5% O2)conditions, compared to related control EC lines. Cells in 6 random high power fields (HPF) were counted, and combined results from 3separate experiments are shown. (C-D) Number of filopodia was assessed using phalloidin staining. (C) Number of filopodia per ECwas quantified and graph shows increased number of filopodia per EC under hypoxic conditions for KO ECs compared to Ctrl. (D) Rep-resentative pictures of phalloidin staining performed on Ctrl and KO ECs under normoxic (21% O2) or hypoxic (0.5% O2) conditions.Data are means ± SEM. *P<0.05.
21% O20.5% O2
10
40
30
20
0
Num
ber
offilo
podi
a/
EC
* *
C
KOH2.1
CtrlH2.1
CtrlH2.2
KOH2.2
KOH2.1

Ctrl H1.1
Ctrl H1.2KO H1.1
KO H1.20.5% O2 Complete media
24hrs 48hrs 72hrs 96hrs
0.5% O2 Serum-free media
24hrs 48hrs 72hrs 96hrs
20
80
60
40
100
0
Via
bilit
y(%
)
Skuli et al., Supplemental Figure 6
Cel
lnum
ber
(.1
06
)
0.5
1
1.5
2
2.5
Day1 Day2 Day3 Day4 Day50
Ctrl H1.1
Ctrl H1.2KO H1.1
KO H1.2
21% O2
Cel
lnum
ber
(.1
06
)
0.5
1
1.5
2
2.5
Day1 Day2 Day3 Day4 Day50
0.5% O2
A
C
B
D
Ctrl H1.1Ctrl H1.2
KO H1.1
KO H1.2*
**
**
* *
* *
*
****
Supplemental Figure 6. HIF-1αα deletion affects endothelial cell proliferation and viability. (A-B) Ctrl and HIF-1α KO lung ECswere incubated under 21% (A) or 0.5% (B) O2 in regular culture media and harvested every 24hrs until the cultures reached confluence.The average cell number of triplicate plates for each condition was determined by counting by hemocytometer following trypsinization.(C-D) Ctrl and HIF-1α KO ECs were cultured at 0.5% O2 in the presence (C) or absence (D) of serum and viable cells were counted atindicated times. Graphs represent the percentage of viable cell obtained from the average of three independent experiments. Data aremeans ± SEM. *P<0.05 and **P<0.01.
100
20
80
60
40
0
Via
bilit
y(%
)
Ctrl H1.1
Ctrl H1.2KO H1.1
KO H1.2

CtrlH1.1
CtrlH1.2
KOH1.2
KOH1.1
0
40
60
*
Num
ber
of
bra
nch
poin
ts/f
ield
80
H
20* **
0
15
20
25
30
Num
ber
of
lum
ens\
fiel
d
10
5
CtrlH1.1
CtrlH1.2
KOH1.2
KOH1.1
** **
G
CtrlH1.1
CtrlH1.2
KOH1.2
KOH1.1
0
20
30
40
50
* *
Tota
lcel
lula
rco
rdle
ngth
(mm
)
21% O20.5% O2
* *10
F
* *
Num
ber
ofin
vade
dce
lls/1
00
Xfiel
d
20
80
60
40
0CtrlH1.1
CtrlH1.2
KOH1.1
KOH1.2
D21% O20.5% O2
Ctrl H1.1
KO H1.1
Ctrl H1.2
KO H1.2
0.5% O2C
20
100
60
40
120
016hrs 24hrs
80
0.5% O2
* *
Ctrl H1.1
Ctrl H1.2KO H1.1
KO H1.2
%of
wound
clo
sure
B
20
60
40
016hrs 24hrs
80 21% O2
Skuli et al., Supplemental Figure 7A 21% O2 0.5% O2
Ctrl H1.1
KO H1.1
Ctrl H1.2
KO H1.2
Supplemental Figure 7. HIF-1αα deletion affects endothelial cell network formation and migration. (A) Migration of Ctrl and HIF-1α KOlung ECs was assessed under normoxic (21% O2) or hypoxic (0.5% O2) conditions at several time points using a scratch wound assay. Repre-sentative photographs shown in (A) were taken 24 hrs after scratch. (B) Percentage of wound closure was determined and was significantlydecreased for HIF-1α KO ECs compared to the control cells at 24 hrs after scratch and under hypoxic (0.5% O2) conditions. (C) EC invasionwas further assessed using a Boyden’s chamber assay. Photographs represent Ctrl and HIF-1α KO EC invasion after 8 hrs under hypoxic (0.5%O2) conditions. (D) Number of invaded cells was quantified at 8 hrs under normoxic (21% O2) or hypoxic (0.5% O2) conditions. Graphs showdecreased invasion for HIF-1α KO ECs compared to control cells at 0.5% O2. (E) EC network/capillary formation was assessed using aMatrigel assay. Photographs represent Ctrl and HIF-1α KO EC capillary formation after 8 hrs under normoxic (21% O2) or hypoxic (0.5% O2)conditions. (F-H) Quantifications of EC network formation reveal significant decrease in total cellular cord length and number of lumens andbranch points for HIF-1α KO ECs compared to control cells at 0.5% O2. Magnification X100. Scale bars: 100 µm. Experiments were performedin triplicates. Data are means ± SEM. *P<0.05, **P<0.01.

50
200
150
100
250
0Num
ber
of
cells
atta
ched
/HPF
Fibronectin
300
CtrlH2.1
KOH2.1
CtrlH2.2
KOH2.2
D
*
Hypoxia 0.5% O2
DMSODAPT
Matrigel
100
250
200
150
300
0Num
ber
of
cells
atta
ched
/HPF
50
CtrlH2.1
KOH2.1
CtrlH2.2
KOH2.2
C
*
*
Hypoxia 0.5% O2
DMSODAPT*
Hey2
0
10
8
Fold
Hypoxi
cIn
duct
ion
4
6
2
CtrlH2.2
KOH2.2
DMSODAPT
**
4
3
Hes1
2
1
0Fold
Hypoxi
cIn
duct
ion
DMSODAPT
CtrlH2.1
KOH2.1
**
B
Skuli et al., Supplemental Figure 8
Ctrl H2.1 KO H2.1
Actin
NICD
Normoxia 21% O2 Hypoxia 0.5% O2
Ctrl H2.1 KO H2.1
DAPT 10µM + + ++- - - -
A
Supplemental Figure 8. Notch pathway inhibition phenocopies HIF-2αα deficiency in endothelial cell adhesion. (A) Western blotanalysis of Notch Intracellular Domain (NICD) protein expression in Ctrl and KO ECs subjected to 21% (Normoxia) or 0.5% (Hypoxia) O2for 6 hrs and treated or not with a Dll4/Notch pathway inhibitor (DAPT). (B) Hypoxic induction of Dll4/Notch pathway target gene expres-sion was assessed by qRT-PCR in Ctrl and KO lung ECs treated with DAPT. The relative ratio of hypoxic to normoxic gene expression(fold hypoxic induction) is shown for control and KO ECs. (C-D) EC adhesion was assessed using Matrigel and Fibronectin adhesionassays. Quantifications of EC adhesion reveal no significant difference in Matrigel (D) or Fibronectin (E) adhesion between untreated KOECs and DAPT-treated control cells at 0.5% O2. Experiments were performed in triplicates. Data are means ± SEM. *P<0.05, **P<0.01.
0.4
0.3
Hes1
0.2
0.1
0Fo
ldH
ypoxi
cIn
duct
ion
DMSODAPT
CtrlH2.2
KOH2.2
**
4
3
Hey2
2
1
0Fold
Hypoxi
cIn
duct
ion
DMSODAPT
CtrlH2.1
KOH2.1
**
Ctrl H2.2 KO H2.2
Normoxia 21% O2 Hypoxia 0.5% O2
Ctrl H2.2 KO H2.2
+ + ++- - - -

A
Skuli et al., Supplemental Figure 9
Figure 9. Dll4 and Dll4/Ang2 are sufficient to reverse endothelial HIF-2αα deletion phenotypes in a hindlimb ischemia model. (A-B) GFPand Dll4 expression were assessed in adductor of Ctrl and KO mice after intramuscular injection of an empty viral vector (expressingGFP), a viral vector expressing Dll4 or Dll4 vector in combination with intravenous Ang2. (A) Representative images showing GFPexpression in muscle fibers and ECs stained with Rhodamine lectin. Arrowheads show ECs expressing GFP. (B) Representative imagesshowing Dll4 expression in muscle fibers and endothelial cells stained with Rhodamine lectin. Arrowheads show ECs expressing Dll4.Please note that the differences in GFP and Dll4 expression patterns, particularly in muscle fibers, may be due to the variable efficiency ofthe viral vector to express either GFP or Dll4 in this tissue and the detection methods. Scale bars: 20 µm. Magnification 400X. (Ctrl n=20,KO n=20).
DAPI GFP Lectin Merged
Ctrl
KO+
empty vector
KO + Dll4
KO + Dll4/Ang2
Ctrl+
empty vector
DAPI Dll4 Lectin MergedB

100
Ctrl KO
20
40
60
80
AmputationNecrosisRecovery
Ani
mal
s(%
)
0
p<0.05
KO+ Dll4
KO+ Dll4/Ang2Empty
vector
p<0.05
p<0.01D
pre post d7 d14 d210
1
2
3
4
Foot
Mov
emen
tSco
re
*
Ctrl + empty vectorKO + empty vector
KO + Dll4/Ang2KO + Dll4
*
**
0
0.2
0.4
Lim
bpe
rfusi
onra
tio(L
aser
Dop
pler
)
Ctrl + empty vectorKO + empty vector
KO + Dll4/Ang2KO + Dll4
Day7
0.6
0.8
1
*
A
0
500
1000
1500
2000
2500
Low
High
Skuli et al., Supplemental Figure 10
Figure 10. Dll4 and Dll4/Ang2 are sufficient to reverse endothelial HIF-2αα deletion phenotypes in a hindlimb ischemia model. Blood flow,foot movement score and necrosis were assessed for Ctrl and KO mice after femoral artery ligation and intramuscular injection of an emptyviral vector, a viral vector expressing Dll4 or Dll4 vector in combination with intravenous Ang2. Blood flow was quantified by laser Doppler.(A-B) Representative images (A) obtained by laser Doppler showing the efficiency of the surgery distally to the occlusion side and the pro-gressive blood flow recovery after 21 days for Ctrl and KO mice. (B) Quantitative laser Doppler analysis showing the left-to-right limb ratioafter occlusion of the femoral artery at day 7. KO mice display a delayed restoration in perfusion compared to Ctrl mice. However, flow iscompletely restored for mice injected with a viral vector expressing Dll4 or Dll4 vector in combination with Ang2. (C) Foot movement wasdetermined and scored between 0 and 4 as a functional read out parameter to assess flow deficits after ischemia. Active foot movementwas significantly impaired in KO mice and restored in the presence of Dll4 vector or Dll4 vector in combination with Ang2. (D) Percentageof mice in each group presenting necrosis or amputation was determined. More KO mice had to be euthanized due to amputation or necro-sis of the limb. Injection of Dll4 vector or Dll4 vector in combination with Ang2 significantly decreases necrosis and amputation. (Ctrl n=20,KO n=20) Data are means ± SEM. *P<0.05 and **P<0.01.
Pre Post Day7 Day14 Day21
Ctrl+
empty vector
KO+
empty vector
KO + Dll4
KO + Dll4/Ang2
B C
3000