Structure of Mutant Human Oncogene Protein Determined
Transcript of Structure of Mutant Human Oncogene Protein Determined
SCIENCE
Structure of Mutant Human Oncogene Protein Determined
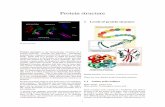
Primary structural differences between normal and transforming human c-Ha-tas oncogene proteins are localized in the loop regions (red) that interact with the β-phosphate ofguanosine diphosphate (tan)
The protein encoded by a mutant human oncogene differs only slightly in structure from the native protein that initiates normal cell division, a finding that may complicate efforts to develop inhibitors of the mutant protein, according to chemists at the University of California, Berkeley.
Sung-Hou Kim, a Berkeley chemistry professor and senior scientist at Lawrence Berkeley Laboratory, last year reported the x-ray structure of the protein encoded by the normal c-Ha-ras gene, a protein believed to signal cells to start or stop dividing through its interaction with guanosine triphosphate (GTP).
Kim has now determined the structure of the protein encoded by a transforming c-Ha-ras oncogene in which a valine codon replaces the normal glycine codon at position 12 in the gene [Nature, 337, page 90 (1989)]. The differences in the structures of the mutant and normal proteins are located primarily in a loop that interacts with the β-phosphate of a bound guanosine diphosphate (GDP) molecule.
Working with Kim at Berkeley were graduate students Liang Tong and Michael V. Milburn, postdoctoral fellow Abraham M. de Vos, and laboratory chemist Jarmila Jancarik. Kim collaborated in the research with Susumu Nishimura and Shigeru Noguchi at the National Cancer Center Research Institute, Tokyo, and Eiko Ohtsuka and Kazunobu Miura at Hokkaido University, Sapporo. The research was supported by the National Institutes of Health; Department of Energy; Japanese Ministry of Health & Welfare; and Merck, Sharp & Dohme.
In a healthy cell, the normal ras protein binds with GTP and transmits signals from the cell membrane to the cell interior that initiate a
cascade of reactions that cause the cell to divide, Kim explains. This ras protein exhibits GTPase activity, and in the normal course of events, the protein cleaves a phosphate ion from the bound GTP to produce GDP, which signals the cell to stop dividing.
The mutant ras protein, which has been found in almost all pancreas tumors and about half of colon tumors as well as tumors from the breast, bladder, and lung, appears to have a significantly reduced GTPase activity. Kim says that the rate of conversion of GTP to GDP in the mutant protein is less than 5% of what it is in the normal protein.
The recently reported research suggests why this might be so. The ras protein contains six β-strands, four α-helices, and nine connecting loops. The protein consists of two structural domains—the amino-
terminal domain, containing the first 75 amino acid residues (including the first three β-strands and one α-helix), which binds phosphate; and the carboxy-terminal domain (including the last three β-strands and three α-helices), which recognizes guanine.
The largest differences in structure were found in the first loop in the amino-terminal half of the molecule, corresponding to amino acid residues nine to 18, Kim says. Taken together, these differences result in a loop that is about 2 A larger in the mutant protein than the corresponding loop in the normal protein. This results in the loss of two hydrogen bonds from the protein backbone amino groups of residues 12 and 13 to the /3-phos-phate of the bound GDP molecule. (The x-ray structure was determined for the protein with a bound GDP.)
Kim had suggested previously that
January 16, 1989 C&EN 31
IBM has the chemical solutions youve been looking for·
The more complex your chemistry problems, the simpler the solution: IBM scientific computing for chemistry.
Featuring a wide variety of IBM systems, specialized applications, development tools, service and technical support, IBM's chemical solutions allow chemists to manage information more effectively and work more productively.
IBM's scientific computing for chemistry combines both IBM's extensive application library and a wide variety of popular, third-party software packages. Together, they cover a vast array of disciplines and operations within the chemical industry: computational chemistry, structural and relational data bases, PC - and host-based technical publishing, laboratory information systems, high-resolution imaging,
IBM and Personal System/2 are registered trademarks of IBM Corporation. © IBM 1989
statistical analysis, and much more. The unparalleled breadth of the IBM product line lets you put the exact amount of
processing power you need exacdy where you need it—from IBM® Personal System/2® computers in the lab, to departmental systems, to the most powerful IBM systems with supercomputing capabilities. Plus, IBM connectivity across the board means that all your systems will work together today, and grow together as your requirements do.
From research to manufacturing, workstations to supercomputers, IBM is your chemical solution.
To find out more, or to have an IBM Marketing Representative contact you, simply call 1-800-IBM-2468, ext. 40.
ORGANIC INTERMEDIATES FROM SWITZERLAND
pharmaceuticals agrochemicals dyestuffs
flavors fragrances photochemicals
Ν - Hydroxyphthalimide
O:;NOH ο
%iv £3
OH
4-Amino- " Υ ~ Ν Ο , 3-nitrophenol NH2
4-Ethylcyclohexanone
υ 0 CH2CH3
2,6-Dimethylpiperidine, cis
2,4-Diamino-1,3,5-trimethyl-benzene (2,4-Diamino-mesitylene)
CHft
H , C ^
NH2
NHL
5-Nitroisophthalic acid and derivatives COOH
A, Custom synthesis against secrecy agreement Our traditional processes: • nitration • catalytic hydrogénation and other reactions
US-Agents: HENLEY CHEMICALS, INC. 50, Chestnut Ridge Road Montvale, NJ 07645 Phone (201)307-0422 FAX (201) 307-0424
^ — 75 years —
R DOTTIKON CH-5605 Dottikon/Switzerland Phone: 057 26 11 55 Telex: 827 923 ssf ch Telefax: 057 24 21 20
CIRCLE 29 ON READER SERVICE CARD 34 January 16, 1989 C&EN
Science
this loop would straddle the phos-phodiester bond between the β- and γ-phosphates of GTP. This bond is the prime candidate to be the catalytic site for GTP hydrolysis in the normal protein. The loss of two hydrogen bonds may alter the orientation of the jS-phosphate group, Kim says, and thus somehow interfere with the hydrolytic reaction.
Working out the complete mechanism of loss of GTPase activity will require determination of the structure of native and mutant ras proteins bound to GTP. This effort is complicated by the proteins' hydrolysis of GTP. Thus the work must be carried out on crystals of the proteins bound to GTP analogs that
contain a bond between the β- and 7-phosphates that is not susceptible to hydrolysis. Efforts to produce crystals of such complexes are under way in Kim's laboratory.
The similarity of structures between the mutant and the normal ras proteins may complicate efforts to develop a drug to deactivate the mutant protein and thus halt tumor growth. "The drug must deactivate the mutant form of the protein without deactivating the normal protein, which is essential to normal cell growth," Kim points out. "It's going to be a difficult job, but it's theoretically possible. Chemists are pretty clever."
Rudy Baum
EDUCATION
Management-of-technology program debuts Satellite broadcasting signals this month are carrying the first courses in a new master of science program initiated by National Technological University in management of technology. NTU plans to have the complete two-year program available in June.
NTU, based in Fort Collins, Colo., is the first electronic university dedicated to nationwide satellite broadcasting of graduate-level advanced engineering and technical education. An accredited, private, nonprofit institution founded in 1984, it is a university without a campus or permanent faculty, and is designed to serve the advanced education needs of busy, mobile engineers, scientists, and technical managers. Ranging from the University of Massachusetts in the Northeast to the University of Arizona in the Southwest, some 28 institutions participate in the network.
So far, NTU's focus has been fairly electrical in nature. The new program brings the number of M.S. programs offered to six. The other five are computer engineering, computer science, electrical engineering, engineering management, and manufacturing systems engineering.
"Management of technology has been identified as one of the top national priorities, especially in view
of the intensity of international competition," says Robert W. DeSio, vice president for development and long-range planning for NTU. "Because management of technology is a relatively new discipline and interdisciplinary in nature, a traditional university would have difficulty in bringing together the required educational programs," he adds.
NTU is in the second phase of development of the new program. The first phase focused on format and curriculum.
Students will be nominated to the program by each of NTU's 60 sponsoring corporations and government agencies. They will be managers who are expected to advance into senior corporate management during their careers. They will have the financial support of their corporations or agencies and will be freed from enough of their regular responsibilities to finish the program in 24 months.
Students will as usual take courses at their work sites through NTU's instructional television network. But in a departure from the format of other NTU programs, they will be brought together during the two-year period for seven one-week intense residencies at different participating university campuses.
James Krieger