Micro - · PDF file... (micro=small, minute) ... single-stranded, circular DNA phage B. Model...
Transcript of Micro - · PDF file... (micro=small, minute) ... single-stranded, circular DNA phage B. Model...

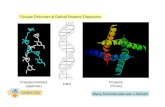
VIROLOGY - MCB 5505 VIRUS FAMILY: MICROVIRIDAE (micro=small, minute) I. DISTINGUISHING CHARACTERISTICS A. Small, single-stranded, circular DNA phage B. Model for circular genomes & plasmids C. Rolling circle DNA replication D. φX174 first DNA genome sequenced (Sanger et al, 1977) E. Extensive overlapping genes (Barrell, 1976) II. STRUCTURE (for φX) A. SIZE: 35 nm in diameter, PW=6.4MD, 114S B. ENVELOPE: NONE 1. GLYCOPROTEINS: 2. OTHER PROTEINS: 3. MATRIX PROTEIN: C. NUCLEOCAPSID 1. NUCLEIC ACID a. TYPE: DNA BALTIMORE TYPE: II b. STRANDED: SS, CIRCULAR c. POLARITY: + d. MOL. WT.: 1.7 MD e. # GENES: 11 2. GENETIC (PHYSICAL) MAP: φX174 5386 N - CIRCULAR A* 1 |------| 5386/1 -|-------------|-----|--------|---|-|--------|-|------|------|- A C D J F G H |---|--| |---| B K E Pa Pb Pd 3. CAPSID a. SYMMETRY: Icosahedral, T=1 b. CAPSOMERS: 12 pentons (5 pF, 5 pG, 1 pH) c. SIZE: 27 nm d. COMPOSITION (1) PROTEINS 60 copies Major capsid (F) protein, 60 kD 60 copies Major Spike (G) protein, 20 kD 12 copies Spike/Pilot (H) protein, 37 kD (2) OTHER COMPONENTS 60 copies Internal (J) protein, 4 kD (DNA binding) 4. NON STRUCTURAL PROTEINS Nickase/ligase (A) protein 60 kD Host DNA turnoff (A*) protein 37 kD Scaffolding (in) (B) protein 30 kD Maturation (C) protein 6 kD Scaffolding (out) (D) protein 14 kD Lysis (E) protein 10 kD Membrane (K) protein 8 kD VIRUS FAMILY: MICROVIRIDAE

III. CLASSIFICATION AND CHARACTERISTIC MEMBERS GENERA PROPERTIES MEMBERS Microvirus Coliphage φX174, G4, α3 Bdellomicrovirus Bdellophage MAC1 Chlamydiamicrovirus Chlamydiaphage Chp-1 Spiromicrovirus Spiroplasma phage SpV4 IV. VIRAL MULTIPLICATION A. ABSORPTION: Spike or pilot protein attaches to specific lipoproteins on surface of E. coli. B. PENETRATION: Pilot protein enters with DNA. C. UNCOATING: During penetration. D. GENE EXPRESSION: The ss, circular DNA genome is converted to a ds DNA, the RF or Replicative Form. E. coli enzymes are responsible. Starts between F & G. Transcription is initiated from several (three) promoters. Proteins are made in different amounts depending on the strength of promoter; mRNAs are polygenic. Circular bidirectional replication proceeds from φX origin (in A gene), pA, a nicking protein is required for this. Later rolling circle replication also proceeds from this site, with pA remaining attached to the 5’-end of single stand. Concatemers are formed. pA nicks and recloses ss circles - new genomes.
E. GENOME REPLICATION: ABOVE - What are RFI, RFII, and RFIII?
F. ASSEMBLY: The capsid self assembles using the
scaffolding protein (pB) which provides a temporary core. An external scaffold is provided by pD. Several precursor particles appear before the entire capsid assembles: 6S, 9S, 12S, 108S & 132S. The mature particle with DNA is 114S.
G. BUDDING AND/OR RELEASE: Particles accumulate in the cell, and if no lysis occurs or is late, more particles accumulate. Lysis is catalyzed by hydrophobic protein, pE.