Cloning and Expression of Exo-β-1,4-Cellobiohydrolase Gene from Bacillus pumilus
Transcript of Cloning and Expression of Exo-β-1,4-Cellobiohydrolase Gene from Bacillus pumilus
Cloning and Expression of Exo-β-1,4-Cellobiohydrolase Gene From Bacillus Pumilus
Meng-jiao Yan1,a, Song Wei1,b, Ya-li Dai1,cand Lin Yuan*,1,d 1Institute of Life Sceince, Inner Mongolia University, Huhhot, China
[email protected], b [email protected], [email protected],[email protected]
Keywords: Exo-β-1,4-cellobiohydrolase, Gene cloning, Expression, Bacillus pumilus
Abstract. The exo-β-1,4-cellobiohydrolase gene(cex) of Bacillus pumilus AC-6 was cloned. The cex
gene from AC-6 is 2100bp, and encodes 700 amino acids. The cex gene has 94% similarities to the
exo-β-1,4-cellobiohydrolase gene sequence from other Bacillus strains. The cex gene was linked with
the vector pGEX-4T-1, and transferred into the competent E. coli BL21 for expression. The result of
protein electrophoresis showed that the protein has expressed, whose molecular weight was about
104kDa. Measured the enzyme activity of the expression protein of the recombinant strains was
5.40U/mL, which is 1.20 times as the original strains.
Intruduction
Cellulose is the most abundant carbohydrate resource in the world. With the increasingly depletion of
non-renewable resources on Earth, it is a great significance to degradate the cellulose by
biotransformation. The exoglucanases is an important member of the cellulase system[1~3]. Now, the
research of cellulase is focus on the gene cloning and expression, site-directed mutagenesis, secretion
and so on[4~6]. We cloned the exo-β-1,4-cellobiohydrolase gene from Bacillus pumilus AC-6, and
expressed it in E. coli.
Materials and Methods
Strains and Plasmids. Bacillus pumilus AC-6 strain was isolated from a chemical fibre factory in
Hohhot, in China. The E.coli DH5α and BL21 were used for gene cloning and expression,
respectively. All E.coli strains were cultivated in Luria-Bertani (LB: 1.0% tryptone, 0.5% yeast
extract, and 1.0% NaCl) medium at 37°C. Ampicillin was added at a final concentration of
100µg/mL. The plasmid pMD19-T was used for gene cloning, and pGEX4T-1 was used as the
expression vector.
Genome DNA extraction.The strain AC-6 was cultured in LB medium at 37°C for 12-16 h with
shaking at 180rpm. The DNA extraction method was described by Wu Qiong[7]. The broth was
centrifuged at 10,000×g for 10 mins. Added 500µL of TE buffer (100mM Tris, 10mM EDTA) and
8µL of lysozyme into the tube with the collected cells. The mixture was incubated at 37°C for 30 mins.
Then added 2µL of proteinase K and 150µL of 10% SDS. The mixture was incubated at 50°C for 45
mins. Added 100µL of 5mol/L NaCl and 80µL of CTAB/NaCl. The mixture was incubated at 65°C
for 15 mins. Next, added 600µL of a mixture of phenol, chloroform and isoamyl alcohol (vol/vol/vol,
24:25:1), and waited for 5 mins, then centrifuged at 13,000×g for 10 mins. The supernatant was
transferred into a new tube, and added the equal volume of isoamylol. The mixture was cooled at
-20°C for 30mins and centrifuged at 12,000×g for 10 mins. Then, added 500µL of 70% ethanol to
precipitate the DNA, and the solution was centrifuged at 12,000×g for 5 mins. The DNA pellet was
washed by 200µL of cold 70% ethanol. After drying, the pellet was suspended in 30µL of 0.1×TE
buffer. The DNA was quantified by electrophoresis using 1% agarose gel.
Exo-β-1,4-cellobiohydrolase gene cloning and sequence analysis. Two primers, F1
[5’-TTGTCAAACAAAGAACGGTTTTTAACGC-3’] and R1 [5’-TTAGTTGATCA
GACGATGATATTCTGCGT-3’], were used for amplification of the cex gene. And two primers
which has the restriction sites sequence, F[5’-GAGGATCCTTGTCAAA CAAAGAACGGT-3’] and
Advanced Materials Research Vol. 647 (2013) pp 150-154Online available since 2013/Jan/11 at www.scientific.net© (2013) Trans Tech Publications, Switzerlanddoi:10.4028/www.scientific.net/AMR.647.150
All rights reserved. No part of contents of this paper may be reproduced or transmitted in any form or by any means without the written permission of TTP,www.ttp.net. (ID: 130.15.241.167, Queen's University, Kingston, Canada-03/10/13,17:00:42)
R [5’-ACGTCGACTTAGTTGATCAGACGATGA-3’], were used to add the restriction sites
sequence to the both sides of the cex gene. The underlined portions in the primer sequences represent
the recognition sites for the restriction enzymes BamHI and SalI, respectively. The PCR reaction was
performed in a 25µL volume, containing 20ng genomic DNA, 0.2mM dNTPs, 1.5mM MgCl2, 0.2µM
of each primer(Transgene,Beijing,China), and 0.5U Taq polymerase (Sangon, Shanghai, China). All
amplifications were performed with an initial denaturing at 95°C for 5 min, followed by 35 cycles of
95°C for 60 s, 54°C for 60 s and 72°C for 90 s, and a final elongation at 72°C for 5 min. Amplified
PCR products were purified by the DNA fragment purification kit (Takara, Dalian, China).The
purified PCR fragment was ligated with the vector pMD19-T, and the ligated product was
transformed into E.coli DH5α competent cells using a standard protocol. Positive transformants were
selected and confirmed by colony PCR. The final validated positive clone of pMD19-T-cex was sent
to Transgene Co. Ltd for sequence determination. The lipase gene sequence was submitted to
GenBank using the BLAST program[8].
Construction of Recombinant Expression Plasmid. Both the cloned cex gene and the plasmid
pGEX-4T-1 were digested with BamHI and SalI, the products were purified and ligated using T4
DNA ligase to form the expression vector pGEX-4T-1-cex. The recombinant expression vector was
transformed into E.coli BL21 competent cells. Positive clones were identified by colony PCR and
double restriction digestion. Detailed protocols were from Sambrook and Russell[9].
Expression of cex gene in E.coli BL21. A transformant of E.coli BL21 harboring pGEX-4T-1-cex
was cultured with vigorous shaking at 37°C overnight in 3mL LB broth containing 100µg/mL
ampicillin. 1ml of the culture was transferred into 100mL fresh LB broth containing the same
concentration of ampicillin. The culture was incubated with shaking at 37°C until the OD600 reached
0.6–0.8. IPTG (final concentration is 0.7mmol/L) was then added into the culture. The induced
protein product was analyzed by SDS-PAGE. Protein was visualized in the gel by Coomassie blue
staining.
The Optimization of Expression. The recombination strain PC211(pGEX-4T-1-cex) was
inoculated as the above. The optimal concentration of IPTG was determined between 100µg/mL and
2000µg/mL. The optimal induction time was determined between 1h and 6h after IPTG addition. The
optimal culture temperature for cellulose was determined between 28°C and 43°C. The induced
protein product was analyzed by SDS-PAGE.
Enzyme Assay.One unit of enzyme (U/mL) is the amount of 1µg of glucose produced from the
cellulose per minute at 50±0.1°C and pH 9.5. The measurement was done according to the Chinese
Industrial Standard (QB2583-2003) [10].
Results and Discussion
Cloning and Sequence Analysis of the cex Gene. A cex-encoding gene was amplified from Bacillus
pumilus AC-6 by PCR, the sequence of the cex gene was determined and submitted to GenBank
(accession no. CP000813).
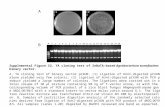
The DNA fragments containing the exo-β-1,4-cellobiohydrolase gene was approximately 2100bp
(Fig.1), which encoded a protein of 700 amino acid residues and its predicted molecular weight was
about 104kDa. The BLAST results showed that the similarity between the cloned gene fragment and
the target gene sequences (Bacillus pumilus SAFR-032 exo-β-1,4-cellobiohydrolase gene) was 94%.
Fig. 1 The cex gene fragments amplified by PCR. Lane M: 250bp DNA Ladder Marker; Lane 1: the
cex gene.
Advanced Materials Research Vol. 647 151
Expression of the cex gene in E. coli BL21. The recombinant plasmid (pGEX-4T-1-cex) was
transformed into E. coli BL21. Expression of the exo-β-1,4-cellobiohydrolase gene was induced by
the addition of IPTG. Protein expression was monitored and analyzed by SDS-PAGE. SDS-PAGE
analysis revealed the presence of a new protein in the supernatant obtained after sonication treatment.
This new protein had an approximate molecular weight of 104 kDa, containing a tag-protein. The size
of the expressed protein agreed well with the predicted size of the
exo-β-1,4-cellobiohydrolase(Fig.2).
Fig. 2 The SDS-PAGE of the recombination protein in E.coli. Lane M: Protein Ruler III marker; Lane
1: empty pGEX-4T-1 vetctor; Lane 3 and 4: empty E.coli BL21(DE3); Lane 5: recombination strain
PC211. The arrow indicates the recombination protein.
The Effects of IPTG Concentration on the Expression of Target Protein. The recombinant
strains were cultured at different IPTG concentrations of 100µg/mL, 200µg/mL, 300µg/mL,
400µg/mL, 500µg/mL, 1000µg/mL, 1500µg/mL, 2000µg/mL. The broth was assayed by the method
of SDS-PAGE. As a result (Fig.3), the target protein can express to the maximum at IPTG
concentrations of 100µg/mL.
Fig. 3 The concentration of IPTG effects on the expression of target proteins. Lane 1: IPTG
concentration is 100µg/mL; lane 2: IPTG concentration is 200µg/mL; lane 3: IPTG concentration is
300µg/mL; lane 4: IPTG concentration is 400µg/mL; lane 5: IPTG concentration is 500µg/mL; lane 6:
IPTG concentration is 1000µg/mL; lane 7: IPTG concentration is 1500µg/mL; lane 8: IPTG
concentration is 2000µg/mL.
The Effects of Induce Time on the Expression of Target Protein. The recombinant strains were
cultured for different times after IPTG addition of 0h, 1h, 2h, 3h, 4h, 5h and 6h. The broth was
assayed by the method of SDS-PAGE. As a result (Fig.4), the target protein can express to the
maximum after induction for 6h.
Figure 4. Effects of induce time on the expression of target proteins. Line 1-7:induction
time:0h,1h,2h,3h,4h,5h,6h
Effect of induce time on the expression of target proteins. Lane 1: empty E.coli BL21; lane 2: induced
time is 6h; lane 3: induced time is 5h; lane 4: induced time is 4h; lane 5: induced time is 3h; lane 6:
induced time is 2h; lane 7: induced time is 1h; lane 8: induced time is 0h.
152 Biomaterial and Bioengineering
The Effects of Temperature on the Expression of Target Protein. The recombinant strains were
cultured at temperatures of 28°C, 31°C, 34°C, 37°C, 40°C and 42°C. The broth was assayed by the
method of SDS-PAGE. As a result (Fig.5), the target protein can express to the maximum at
temperature of 28°C.
Fig. 5 Effect of temperature on the expression of target proteins. Lane 1: induced temperature is 28°C;
lane 2: induced temperature is 31°C; lane 3: induced temperature is 34°C; lane 4: induced
temperature is 37°C; lane 5: induced temperature is 40°C; lane 6: induced temperature is 42°C.
Conclusion
The exo-β-1,4-cellobiohydrolase gene from Bacillus pumilus AC-6 has successfully expressed in E.
coli. Through the optimization of expression conditions, we have found that the recombinant strains
will express the target proteins to the maximum after an induced expression of 6h at 28℃. In addition,
the exo-β-1,4-cellobiohydrolase activity of recombinant strains is 1.20 times higher than that of the
original strain AC-6 by using DNS. Because of the lower enzyme activity of the original strain and
the engineering strain, it will be restricted in the industrial application. So we need to look for a better
expression system, for example, yeast expression system, in order to achieve higher expression and
activity.
Acknowledgement
This research was supported by “field rank college students training plan of the creation and
innovation fund” of Inner Mongolia university.
References
[1] Ding Xinli, Wang Tianhong, Study of the expressions of cellulase from Trichoderma reesei in
Saccharomyces cerevisiae, Liguor-making Science & Technology. 9 (2005)28-35.
[2] GaoFengju, Chen Hui, Wu Qi, Selection and identification of cellulase-producing Bacillus C-36,
Journal of Sichuan Agricultural University. 24 (2006)175-177.
[3] Jing Yinghui, Liu Xiangmei, Chen Shicheng, Screening of an alkali-tolerant β-glycanases
producing strain and its optimization of their enzyme production, Chin. J. Appl. Environ. Biol. 5
(1999) 404-410.
[4] WU Ruiqing, DU Junqi, LI Ming, Deinking technology for mixed wastepaper with highly
efficient deinking agent, Modern Chemical Industry. 30 (2010) 56-60.
[5] JI Yachao, YANGYaru, WANG Zhonglin, LEI Dengying, WEI Yongqin, ZHANG Xiangming,
Pilot test on application of enzymatic deinking agent in ONP deinking line, Paper Chemicals. 22
(2010) 28-30.
[6] Wang Zhonglin, Yang Yaru, Zhang Xiangming, Xie Bifeng, Li Jiang, Lei Yichao, Studies on the
Bio-enzymatic deinking of old newsprint, Paper Chemicals. 41 (2010) 58-60.
Advanced Materials Research Vol. 647 153
[7] Wu Qiong, Yuan Lin, Lu Fuping, Shen Minghua, Liu Chunyan, Bai Yuchen, Screening and
identification of an alkaline cellulase-producing strain, Biotechnology Bulletin. 9 (2010)
205-209.
[8] Chenyan Zhou, Jianyu Bai, Shanshan Den, Cloning of a xylanase gene from Aspergillu susamii
and its expression in Escherichia coli, Bioresource Technology. 99 (2008) 831–838.
[9] J Sambrook, DW Russell. Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor
Laboratory Press, New York, 2001.
[10] CSIC-QB, “QB2583-2003,” China standard press, 2003.
154 Biomaterial and Bioengineering
Biomaterial and Bioengineering 10.4028/www.scientific.net/AMR.647 Cloning and Expression of Exo-β-1,4-Cellobiohydrolase Gene from Bacillus pumilus 10.4028/www.scientific.net/AMR.647.150