INF5410 Array signal processing INF5410 Array signal processing ...
Chapter 31 Transcription and RNA processingpersonal.tcu.edu/yryu/50133/Transcription.pdf ·...
Transcript of Chapter 31 Transcription and RNA processingpersonal.tcu.edu/yryu/50133/Transcription.pdf ·...

Chapter 31
Transcription and RNA processing

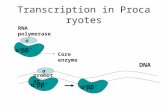
RNA polymerase (RNAP)

E. coli promoters

Components of E. coli RNA Polymerase Holoenzyme (α2ββ'ωσ)

Structure of prokaryotic RNAP

The closed and open state of RNAP
The closed state The open state

Chain elongation

Transcriptional termination in prokaryotes
• Rho-independent termination– Terminator sequence: Hairpin structure with
oligo(U) tail• Rho-dependent termination
– No obvious sequence similarity– Rho factor: Helicase and NTPase activity

Terminator
Poly(U) tail

Eukaryotic RNA polymerases
• RNA polymerase I (RNAP I, RNAP A) - rRNA• RNA polymerase II (RNAP II, RNAP B) - mRNA• RNA polymerase III (RNAP III, RNAP C) - tRNA,
5S rRNA, small nuclear and cytosolic RNAs

RNA polymerase subunits

RNAP II elongation complex
Roger Konberg (2001)

Eukaryotic promoters
• Mammalian RNA polymerase I has a bipartite promoter– Core promoter elements (-31 to +6)– Upstream promoter elements (-187 to -107)
• RNA polymerase II promoters– GC box (Constitutive housekeeping genes)– TATA box (-25 to -30, cell type-specific genes)– CCAAT box (-70 to -90)– Enhancers (selective gene expression)
• RNA polymerase III promoters– Internal and/or upstream promoters

Eukaryotic transcription factors
• At least six general transcription factors(GTFs) – Equivalent of prokaryotic σ factor
• GTFs form preinitiation complex (PIC) along with RNAP II and promoter DNA

Preinitiation complex (PIC)

Control of transcription in prokaryotes
• Efficiency of promoters• Different σ factors• Repressor • Catabolite repression – gene activation • Attenuation• Stringent response

The expression of the lac operon
(Natural inducer)
(Synthetic inducer)

Catabolite repression
• Adequate amount of glucose prevent the full expression of gene specifying proteins involving in the fermentation of numerous other catabolite, including lactose, arabinose, and galactose
• cAMP levels are low in the presence of glucose but rise when glucose become scarse
• Catabolite gene activator protein (CAP)-cAMP complex enhances the transcription of catabolite repressed operons

E. coli araBAD operon

Attenuation

Operons subject to attenuation

Riboswitches• Messenger RNAs that control gene expression through their ability to
bind small molecule metabolites directly • Regulate biosynthesis of thiamine pyrophosphate, vitamine B12,
riboflavin, S-adenosylmethionine (SAM), guanine, adenine, lysine and so on
TPP-dependent mRNA conformations

Stringent response
• In stringent or poor growth conditions, cellular resources are diverted from growth and division to amino acid synthesis
• When there are no charged tRNA's available to bind to the ribosome, RelA synthesizes pppGppand several ribosomal proteins convert pppGppto ppGpp
• (p)ppGpp inhibits a number of energy-consuming cellular processes, including replication and transcription

Posttranscriptional processing
• Eukaryotic mRNA processing– 5’-Cap: 7-methylguanosine (m7G) and 2’-O-methylation– Poly(A) tails– Splicing
• rRNA processing– Prokaryotic rRNA: cleavage of primary transcripts by several
different RNases (and a few methylation)– Eukaryotic rRNA:
• Ribose and base methylation, and subsequent cleavage by RNases• Self-splicing rRNA: group I and II introns
• tRNA processing– 5’-Cleavage by RNAse P– 3’-Cleavage and trimming by many RNAse enzymes– Attachment of 3’-CCA to eukaryotic tRNA primary transcripts– Modified bases

5’-Cap of eukaryotic mRNA

Processing of pre-mRNA or heterogeneous nuclear RNAs (hnRNAs)

The splicing reaction

Spliceosome
• Splicing takes place in the spliceosome
• Each spliceosome is composed of five small nuclear RNA proteins, called snRNPs, and other protein factors

Alternative splicing: Multiple proteins from a single gene
Alternative splicing in the rat α-topomyosin gene

• Silencing of gene expression, triggered by the presence of double-stranded RNA homologous to portions of the gene
• Dicer cleaves the long double-stranded RNAsinto short 21- to 25-base-pair small interfering RNAs (siRNAs)
• siRNAs and several proteins form RNA-induced silencing complex (RISC)
• Unzipping of double stranded siRNA activates RISC, which can in turn bind to the target mRNA and cleave it
RNA interferance (RNAi)

RNA interferance (RNAi)

Posttrancriptional processing of E. coli rRNA

Self-splicing rRNA
Self-splicing rRNA (Group I intron) of Tetrahymena thermophila
2

Small nucleolar RNAs (snoRNAs)
• snoRNAs guide the methylation or pseudouridylation of eukaryotic rRNAs by complimentary base pairing– C/D box: 2’-O-methylation– H/ACA box: peudouridylation
• snoRNAs and other nucleolar proteins form small nucleolar ribonucleoprotein (snoRNP)

tRNA processing
• Cleavage of 5’-end by RNAse P, whose RNA component has the catalytic function
• Many different RNaseenzymes are involved in 3’-cleavage (or trimming) of prokaryotic initial tRNAtranscripts
• The –CCA ends of eukaryotic tRNAs are posttranscriptionallyappended

Processing of eukaryotic tRNA
Removal of 5’-extension by RNAse P
Removal of intervening sequence (intron)
3’-CCA addition
Modified bases