Supplementary data 4-1-11 - Journal of Bacteriology · 6/9/2011 · Supplementary data Fig....
Transcript of Supplementary data 4-1-11 - Journal of Bacteriology · 6/9/2011 · Supplementary data Fig....

Supplementary data
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
cell free protein lysates from WT M. tuberculosis (Rv) together with various known
concentrations of purified CrgA were resolved on a 15% NuPAGE gel, transferred to
PVDF membrane and probed with α-CrgA antibodies. CrgA bands were quantitated
by QuantityOne software. A standard curve was prepared using pure protein
standards and used to determine the CrgA levels in the lysates. Arrowhead – CrgA
protein.

Fig. S2. (i) FtsZ levels in M. tuberculosis crgA strains. WT, crgA overexpression and crgA
antisense strains were grown for 48 hrs with 100 ng/ml anhydrotetracycline and cells
processed for immunoblotting using indicated antibodies. (ii) CrgA levels do not change
upon FtsZ depletion. M. smegmatis ∆ftsZ, Pami::ftsZ, Ptet::ecfp-crgA was grown with100
ng/ml anhydrotetracycline and without and with 0.2% acetamide for 6 hrs. Bacteria were
pelleted by centrifugation and analyzed by immunoblotting using antibodies to indicated
proteins.

Fig. S3. (A) FtsI and FtsZ levels in M. smegmatis WT and crgA strains expressing
Pami::gfp-ftsI. M. smegmatis strains were grown as described in the text and processed for
immunoblotting. Blots were probed with antibodies to the indicated proteins as described in
the text. FtsI and FtsI-GFP were probed with α-FtsI antibodies. - CrgA overproduction
strain; Δ crgA – crgA mutant strain. (B) Complementation of crgA mutant. M. smegmatis
WT (i), Δ crgA (ii) and Δ crgA PcrgA::crgA (iii) and Δ crgA Pet::ecfp-crgA (iv and v) were
grown in 7H9-AD and examined by brightfield (i - iv) and fluorescence (v) microscopy. Δ
crgA Pet::ecfp-crgA was grown with 100 ng/ml anhydrotetracycline. Black arrowheads –
swollen cells. White arrowheads – ECFP-CrgA localization at midcell or quarter cell
positions. White arrows – polar localization of ECFP-CrgA. Bar – 5 µms.

Fig. S4: (A) CrgA interactions with PBPB, PBPA and FtsQ are specific: crgA, pbpA, ftsQ,
clpX and rodA fusions to the T25 or T18 fragments of adenylate cyclase were cloned in
various BACTH vectors (Table 1) and interactions examined as described in Fig. 4A legend.
E. coli BTH101 recombinants bearing indicated combinations of plasmids were plated on
LB agar containing IPTG and X-gal. Blue colonies indicate strong positive interactions

(Inset). RodA-PbpA is a positive interaction. Mean ± SD from 3 independent experiments
are shown. (B) (i). Tag-free FtsZ and CrgA do not bind non-specifically to cobalt affinity
resin. Tag-free purified FtsZ and CrgA were mixed together and allowed to bind cobalt
affinity resin and bound proteins eluted as described for Fig. 4B (i). Load (L), wash (W) and
elution (E) fractions were resolved on SDS-PA followed by immunoblotting with indicated
antibodies. (ii). N-terminal 1-51 aa of CrgA are sufficient for interaction with FtsZ. Purified
His-FtsZ was mixed with purified 1-51 aa CrgA protein and pull-down assay performed
using cobalt affinity resin as described for Fig. 4A. Load (L), wash (W) and elution (E)
fractions were resolved on SDS-PA followed by immunoblotting with indicated antibodies.

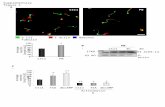
Fig. S5. (A) N-terminal 1-51 aa are sufficient for localization of CrgA to the septal sites.
ECFP-CrgA or ECFP-∆C-term-CrgA fusion was localized in wild type M. smegmatis as
described under Fig. 3A. CrgA domains cartoon: I and O are cytoplasmic and
extracytoplasmic regions; T1 and T2 are 1st and 2nd transmembrane domain. Arrowhead –
midcell localization. Bar – 5 µm. (B) Bacterial 2-hybrid assays with truncated FtsI and CrgA
proteins. Shown BACTH assays were carried out as described for Fig. 4A. See table 1 for
plasmid descriptions. FtsI679 is full-length FtsI. FtsI124 and FtsI298 are FtsI N-terminal 1-
124 and 1-298 aa proteins, respectively. CrgA1-51aa is CrgA N-terminal 1-51 aa protein.
GCN4-GCN4 is positive control.

Fig. S6. Reverse-phase HPLC profile of peptidoglycan from WT or CrgA overproducing M.
smegmatis strains. Cells from overnight cultures of both strains were mechanically
disrupted, boiled in SDS and digested with proteases and muramidases as described
(Lavollay et al. 2008). Muropeptides were reduced with Sodium borohydrate and injected
onto C18 nucleosil column and separated in H2O+TFA with a Acetonitrile Gradient (0-20%)
(Table S1; see also Lavollay et al. 2008 for more details).
Table S1: Muropeptide composition
A Tripeptide
B Tetrapeptide -GlcNac
C Tetrapeptide
D Peptide contaminant m=522 (always here)
E Tri-Tetra -GlcNAc 3-3 crosslink (L,D-transpeptidation)
F Tri-Tri 3-3 crosslink (L,D-transpeptidation)
G Tri-Tetra 3-3 crosslink (L,D-transpeptidation)
H Tetra-Tetra 4-3 crosslink (D,D-transpeptidation)
I Peptide contaminant m=679 (always here)
J Anhydro Tri-Tetra 3-3 crosslink (L,D-transpeptidation)

Table S2: Oligonucleotide used in the study Oligo Name Sequence (5’-> 3’) Plasmid/ gene
Reference
CrgA-PacI AGAACCTTAATTAAGAGCCCCACCAGGGAGGAA GCCGAACGATGCCCAAGTCCAAGGTCCG
forward for crgA in pDS5 and pPP91
CrgA-SwaI ATCGGATTTAAATATCAATGCCAGCGCATCGTGA reverse for crgA pDS5, ecfp fusion in pPP93
CFP-PacI F AACCTTAATTAAGAGCCCCCACCAGGGAGGAAG CCGAACGATGGTGAGCAAGGGCGAGGA
forward for ecfp
CFP-BamHI CGGGATCCCTGCAGGTTGTTGTTCTTGTACAGCT CGTCCATG
reverse ecfp

CrgA-BamHI GCGGATCCATGCCCAAGTCCAAGGTCCG forward for crgA in pMAL and ecfp fusion
CrgA-end-HindIII
GAGCCCAAGCTTTCATCAATGCCAGCGCATCGTGA reverse for crgA in pMAL
aCrgA-PacI AGAACCTTAATTAAGAGCCCCACCAGGGAGGAA GCCGAACGTCAATGCCAGCGCATCGTGA
forward for crgAMtb antisense
aCrgA-SwaI ATCGGATTTAAATATGCCCAAGTCCAAGGTCCG reverse for crgAMtb antisense
CrgA-BamHI TCTAGAGGATCCCATGCCCAAGTCCAAGGTCCGC forward for crgA BACTH
CrgA-KpnI TTACTTAGGTACCCGATGCCAGCGCATCGTGAGCA reverse for crgA BACTH
Yfp XbaI TATAGTCTAGATTATTACTTGTACAGCTCGTCCA reverse for ftsZ-yfp

FtsZ-NdeI CAGCCATATGACCCCCCCGCACAACTACC forward for ftsZ-yfp
PBPA- BamHI
TCTAGAGGATCCCATGAACGCCTCTCTGCGCCGA forward for pbpA BACTH
PBPA-KpnI TTACTTAGGTACCCGTGGTTCCCCCTGCAGTGCGGC reverse for pbpA BACTH
RodA-XbaI TTGCTCTAGAGATGACGACACGACTGCAAGC forward for rodA BACTH
RodA-KpnI TTACTTAGGTACCCGTACGCGCCTGATGACCTCGG reverse for rodA BACTH
FtsI-BamHI GCGGATCCGTGAGCCGCGCCGCCCCCAG forward for gfp-ftsI
FtsI298-XbaI
TTGCTCTAGATTACTATTGGCCGGCCGCCTGCGCACG reverse for ftsI BACTH
FtsI 124 XbaI
TTGCTCTAGATTACTAGTAGCTGCCGGGGATGACGAC reverse for ftsI BACTH
FtsI XbaI TTGCTCTAGATTACTAGGTGGCCTGCAAGACCAA reverse for gfp ftsI

FtsI NdeI GGAATTCCATATGAGCCGCGCCGCCCCCAGGC forward for ftsI pET-19b
FtsI BamHI CGCGGATCCCTAGGTGGCCTGCAAGACCAA reverse for ftsI pET-19b
CrgA 51aa SwaI
TCGGATTTAAATTCAAAACACCATTAACCAGATGAG reverse for crgA pPP91
CrgA 27aa PacI
CACTGGATCCATGGGACCGTCGAGCGTATGGTT forward for crgA pPP93

Supplementary methods
Intracellular levels of CrgA in M. tuberculosis. Intracellular lysates of M.
tuberculosis strains were prepared and the CrgA levels were quantitated by immunoblotting
and normalized to SigA as described (Dziedzic et al. 2010). Cell lysates were resolved on
NuPAGE polyacrylamide gels, transferred to PVDF membrane and probed with anti-CrgA
antibodies diluted to 1: 25,000. Immunoblots were processed with the ECF Western blotting
kit (GE life sciences, Piscataway, NJ) and scanned on a Bio-Rad Molecular Imager. For
quantitative immunoblotting, known amounts of purified CrgA protein were quantified by
volume analysis function of the QuantityOne software and standard curves were plotted.
Lysates loaded on the same gel as the standards were then quantitated using the standard
curve as described previously (Dziedzic et al. 2010). Concentration of purified CrgA protein
was determined using the BCA protein assay kit (Pierce). The number of colony forming
units per ml culture at appropriate OD600 was used to calculate the number of CrgA molecules
per cell. The calculated intracellular CrgA levels are average from lysates derived from 3
independent cultures of M. tuberculosis.