NUANCE, a giant protein connecting the nucleus and actin … · 2002. 7. 2. · repeats, similar to...
Transcript of NUANCE, a giant protein connecting the nucleus and actin … · 2002. 7. 2. · repeats, similar to...

IntroductionMany biological processes depend on the balance between thedynamics and the stability of the actin cytoskeleton. Thecellular organization of the microfilament system is thereforecharacterized by a large variability and flexibility. Proteins ofthe α-actinin superfamily utilize a double calponin homologydomain to arrange actin filaments in bundles or meshworks andto link them to the plasma membrane (Matsudaira, 1994). Themodular organization of the proteins of the α-actininsuperfamily (Puius et al., 1998) allows the building ofextremely long protein chains. The largest proteins mainlyrepresent two families: plakins (plectin and dystonin) (Brownet al., 1995; Wiche, 1998) and spectrins (dystrophin, utrophinand spectrin itself) (Ahn and Kunkel, 1993; Tinsley et al.,1992). Plakins are characterized by a tripartite structure, wherea central coiled-coil rod separates two globular domains: theN-terminal plakin domain and the C-terminal domain, whichis composed of plectin repeats (Ruhrberg and Watt, 1997). Thecharacteristic feature of spectrins is the rod domain, which iscomposed of multiple triple helical spectrin repeats (Pascual etal., 1997).
The identification of the Drosophila protein kakapo (alsoknown as groovin) (Gregory and Brown, 1998; Prokop et al.,1998; Strumpf and Volk, 1998) and its mammalian homologueMACF (also described under the names trabeculin-α andmacrophin) (Leung et al., 1999; Okuda et al., 1999; Sun et al.,1999), as well as BPAG1-a and BPAG1-b forms of theBPAG/dystoninlocus (Leung et al., 2001b), brought confusionto this classification, since these proteins, which are larger thanthe other known plakin and spectrin proteins, harbor bothdomains found in plakin and spectrin and may represent afusion product of two precursor genes. An alternativehypothesis proposes that the ancestral gene resembled the
shortstop/kakapogenes of Drosophila and Caenorhabditis(Leung et al., 2001a). Owing to their multiple binding sites formicrotubules, intermediate filaments and microfilaments, thesegiant proteins are recognized as cytolinkers, integrating thecytoplasmic cytoskeleton with membranes and submembranecomplexes (Fuchs and Yang, 1999; Karakesisoglou et al.,2000).
The recently described protein calmin, which also possessesa N-terminal actin-binding domain (ABD) of the α-actinintype, does not fall into any of the above categories, as it doesnot harbor either a coiled-coil rod domain or any other commonmotifs (Ishisaki et al., 2001). Instead it has a C-terminaltransmembrane domain (TMD) that targets cytoplasmicreticular structures and therefore represents the first integralmembrane α-actinin-related protein known so far. However,some transcripts of calmin are differentially spliced and lackthe TMD. The distribution pattern of the calmin isoforms didnot overlap with that of the actin cytoskeleton. Here we reportthe molecular characterization and cellular localization ofNUANCE, the largest known protein of the α-actininsuperfamily. The 796 kDa protein harbors multiple dystrophin-like spectrin repeats but lacks domains characteristic forplakins. Hence, it is related to the spectrin family of the α-actinin superfamily. Unlike other related proteins, NUANCE isassociated with the nuclear envelope (NE) via the predictedTMD. This enormously large protein may harbor numerousbinding sites and serve as a scaffold for the assembly of variousprotein complexes.
Materials and MethodsMolecular cloningThe clones AA480953 and AA761297, containing partial NUANCE
3207
NUANCE (NUcleus and ActiN Connecting Element) wasidentified as a novel protein with an α-actinin-like actin-binding domain. A human 21.8 kb cDNA of NUANCEspreads over 373 kb on chromosome 14q22.1-q22.3. ThecDNA sequence predicts a 796 kDa protein with an N-terminal actin-binding domain, a central coiled-coil roddomain and a predicted C-terminal transmembranedomain. High levels of NUANCE mRNA were detected inthe kidney, liver, stomach, placenta, spleen, lymphaticnodes and peripheral blood lymphocytes. At the subcellularlevel NUANCE is present predominantly at the outer
nuclear membrane and in the nucleoplasm. Domainanalysis shows that the actin-binding domain binds to F-actin in vitro and colocalizes with the actin cytoskeletonin vivo as a GFP-fusion protein. The C-terminaltransmembrane domain is responsible for the targeting thenuclear envelope. Thus, NUANCE is the first α-actinin-related protein that has the potential to link themicrofilament system with the nucleus.
Key words: CH domain, Spectrin repeats, Klarsicht-like domain,Nuclear envelope
Summary
NUANCE, a giant protein connecting the nucleus andactin cytoskeletonYen-Yi Zhen, Thorsten Libotte, Martina Munck, Angelika A. Noegel and Elena KorenbaumInstitute for Biochemistry, Medical Faculty, University of Cologne, 50931 Cologne, GermanyAuthor for correspondence (e-mail: [email protected])
Accepted 1 May 2002Journal of Cell Science 115, 3207-3222 (2002) © The Company of Biologists Ltd
Research Article

3208
cDNA, were obtained from RZPD (Resource Center/PrimaryDatabase of German Human Genome Project) and fully sequenced.The full-length cDNA of NUANCE was cloned by using SMARTRACE cDNA amplification kit (Clontech) and Advantage 2polymerase (Clontech) with mRNA from Burkitt’s lymphoma cellsBL-60 as a template. With 5′-RACE-PCR we extended the cDNA tothe putative 5′ end (fragment 1, Fig. 1A). Several rounds of 3′-RACE-PCR extended the cDNA to a total length of 6.1 kb (fragments from2 to 5, Fig. 1A). Close inspection of the publicly accessible genomicsequence (GenBank) downstream of the identified NUANCE partialcDNA revealed a polyadenylated cDNA of DKFZp434G173(accession number AL080133), which encoded several spectrinrepeats, similar to those found in NUANCE. This suggested that theamplified 6.1 kb fragment of NUANCE and DKFZp434G173 cDNAsrepresent the 5′ and 3′ end of the same giant transcript. The ESTsmatching the genomic DNA from the gap region, which encodedpeptides with homology to dystrophin-like spectrin repeats, were usedto design supplementary primers. The PCR products were cloned intothe pGEM T-Easy vector (Promega) and sequenced. 22 clonescontaining partial NUANCE cDNAs were used to assemble full-length NUANCE cDNA (accession number AF435011). The cDNAof the short ABD-S isoform has been deposited under accessionnumber AF435010. Partial mouse NUANCE cDNAs from RT-PCR ofkidney RNA with the primers designed from the sequence of ESTclones AI747790 and AA498987 were deposited under accessionnumber AF435012.
Expression analyses The expression profile of human NUANCE was studied by probing ahuman multiple tissue expression array (Clontech). The cDNA probeencompassing nucleotides from 247 to 1443, which corresponds tothe NUANCE ABD with the following spectrin repeat, was 32P-labeled using a random prime labeling kit (Stratagene). As a control,the blot was stripped and reprobed with ubiquitin.
Production of the recombinant proteins in E. coli andgeneration of the monoclonal antibodiesDNA fragments encoding the first 285 (6×His-ABD) and 459 aminoacid residues (6×His-ABD-l) were obtained by PCR using fragment2 as a template and inserted into pQE-30 (Qiagen). Recombinantproteins were induced by IPTG in E. coli M15[pREP4] and purifiedusing a Ni-NTA agarose column according to the manufacturer(Qiagen). mAb K20-478 was produced by immunizing mice with thepurified 6×His-ABD-l protein with ImmunEasy mouse adjuvant(Qiagen) as described earlier (Olski et al., 2001).
Actin-binding assayThe F-actin co-sedimentation assay and the quantification of 6×His-ABD bound to F-actin was performed as described previously (Olskiet al., 2001). For high- and low-speed centrifugation assays, sampleswere pelleted at 125,000 g and at 20,000 g, respectively. Actinpolymerization was measured by recording the changes influorescence intensity of pyrene-labeled α-actin monomers asdescribed previously (Korenbaum et al., 1998). The fluorescencemeasurements were made using a Fluoroskan Ascent FL plate reader(Labsystems). Polymerization of 8 µM actin alone or in the presenceof various amounts of the ABD was initiated by addition of 2 mMMgCl2 and 100 mM KCl.
Plasmid construction and transient transfectionsThe constructs GFP-ABD (residues 1-296) and GFP-ABD-S (residues1-262 with additional amino acids AYKN from exon 8a) wereamplified from fragment 2 and 2a, which code for spliced variants,
using primers with extensions for BamHI and KpnI sites and clonedinto BglII/KpnI-cut pEGFP-C1 (Clontech). For the GFP-ABDsr1-2construct (residues 1-531), fragment 2 was digested with BamHI andEcoRI then ligated into BglII/EcoRI-cut pEGFP-C1 vector. For theGFP-sr15-21 (residues 5727-6596), the fragment 13a was excisedutilizing the EcoRI sites of pGEM T-Easy (Promega) and ligated intoEcoRI site of pEGFP-C1. For the GFP-Cterm1 construct (residues6571-6885), the cDNA was amplified from clone 13 and ligated intoEcoRI-cut pEGFP-C2 vector. For the GFP-Cterm2 constructs, clone13b was digested with SacI and EcoRI and inserted into correspondingsites of pEGFP-C1. The GFP-Cterm2∆tm (residues 6642-6834 withadditional 14 amino acids from the extended exon 111) lacking theputative TMD was generated from the GFP-Cterm2 by PstI digestionand religation. To generate GFP-NUA∆460-6643, the GFP-ABDsr1-2was cut with KpnI and BamHI and ligated with the insert of the GFP-Cterm1 (residues 6643-6885), which was amplified using the primerswith added-on KpnI and BglII sites. COS7 cells were transfected byelectroporation. The expression of the fusion proteins was controlledby western blotting using a GFP-specific mAb.
Cell cultureCOS7 cells were grown in DME medium supplemented with 10%FBS (Sigma), 2 mM glutamine, penicillin and streptomycin. For thepropagation of human embryonic kidney cells (293) pyruvate wasadded to the medium. Human T lymphocytes (Jurkat), Burkitt’slymphoma cells BL-60 and B-JAB were maintained in RPMI 1640containing 10% FBS, 2 mM L-glutamine, penicillin and streptomycin.For Latrunculin A (Biomol) treatment, 70%-confluent COS-7 cellswere fixed 15, 30 and 60 minutes after exposure to 1 µM LatA. Therecovery was evaluated by culturing the cells in LatA-free mediumfor another 30, 60 and 90 minutes. For microtubule depolymerizationthe cells were incubated in medium containing 1 µM vincristine(Sigma) or 5 µg/ml colchicine (Fluka) for 4 hours before processingfor immunofluorescence. To promote the formation of lamellipodia,confluent serum-starved monolayers of COS7 cells were wounded byscraping away cells with a P1000 pipet tip. The coverslips were thenincubated in complete media for 6 hours prior to fixation.
ImmunofluorescenceAdherent cells were allowed to attach onto glass coverslips for 1-16hours, rinsed with PBS, fixed in 3% paraformaldehyde andpermeabilized with 0.5% Triton X-100 in PBS for 5 minutes.Alternatively, cells were fixed with methanol at –20°C for 10 minutes.Generally, no difference was observed between the two fixationprotocols except for the cells transfected with the GFP-Cterm2construct. All figures display cells fixed with paraformaldehyde unlessstated otherwise. For permeabilizing with digitonin, fixed cells (3%-paraformaldehyde) were washed in ice-cold PBS and treated with 40µg/ml digitonin (Sigma) in PBS for 5 minutes on ice. For poorlyadherent cells, cover slips were pretreated with poly-L-lysine (1mg/ml), and cells were allowed to attach for at least 3 hours. Cellswere incubated with mAbs against vinculin (Sigma), annexin A7(Selbert et al., 1995), with the maD mAb, which is specific for β-COP(Pepperkok et al., 1993), with JOL2, which is specific for lamin A/C,with LN43, which is specific for anti-lamin B2 (kind gift from FransRamaekers and Jos Broers) or with polyclonal antibodies againstNup358 (kind gift from Elias Coutavas and Günter Blobel) andagainst NO38/B23 (kind gift from M. Schmidt-Zachmann). Cellswashed with PBS were incubated with the appropriate secondaryantibodies conjugated to Cy3 (Sigma), Alexa488 or Alexa568(Molecular Probes) and mounted in Gelvatol/DABCO (Sigma). F-actin was detected with TRITC-labeled phalloidin; for nuclearstaining the DNA-specific dye DAPI (Sigma) was used. Samples wereanalyzed by wide-field fluorescence microscopy (DMR, Leica) orconfocal laser scanning microscopy (TCS-SP, Leica).
Journal of Cell Science 115 (15)

3209An actin-binding protein at the nuclear envelope
Cell fractionation and immunoblotsCOS7 cells were harvested and washed in PBS, homogenized in lysisbuffer (1% SDS, 1 mM sodium vanadate and 100 mM Tris-HCl, pH7.4), incubated at 95°C for 5 minutes and mixed with the proteinsample buffer (Laemmli, 1970) and heated again for 5 minutes to95°C. Proteins were separated on 3-15% gradient SDS-PAGE thentransferred onto PDVF membrane (Millipore). The membraneswere treated with mAb K20-478 followed by enhancedchemiluminescence.
For nuclei preparation, COS7 cells were sonicated in hypotonicbuffer (10 mM HEPES, pH 7.5, 1.5 mM MgCl2, 1.5 mM KCl, 0.5mM DTT, 0.2 mM PMSF) supplemented with Protease InhibitorCocktail CompleteTM, Mini (Boehringer Mannheim). Nuclei weresedimented at 1,000 g at 4°C for 15 minutes. For further fractionationthe supernatant was centrifuged at 100,000 g for 30 minutes at 4°C.Both pellets were resuspended in hypotonic buffer and analyzed onimmunoblots with anti-NUANCE, anti-lamin B2 (LN43) and anti-annexin A7 mAbs.
Computer programsFor alignment of cDNA sequences and database mining, the GCGsoftware package and the BLAST (NCBI) program were used. Proteinsequences were aligned using the programs ClustalW and TreeView.Motif predictions and pattern searches were performed with theExPaSY (SIB) software package.
ResultsCloning and analysis of NUANCE cDNAWe searched for new proteins with homology to the ABD ofα-actinin by screening the human EST database using thepeptide sequence of ABDs of known α-actinin-related proteinsas a query. Two overlapping ESTs, AA480953 and AA761297,were identified that contained a partial cDNA encoding for anovel protein with an ABD that was highly homologous toMACF (Leung et al., 1999) and plectin (Wiche et al., 1991).Both clones were derived from a cDNA library prepared fromhuman tonsillar cells enriched for germinal center B cells.Combining RACE-PCR with the analysis of data availablefrom human non-redundant EST and human genomicGenBank databases, we cloned 21.8 kb of NUANCE cDNA(see Materials and Methods; Fig. 1A). The putative ATG startcodon at position 205 together with the surrounding sequenceAGAATGG matched the Kozak consensus (Kozak, 1987). Anin-frame stop codon was located 26 bp upstream, making thisATG the translational start site. The 3′ untranslated region(3′UTR) of 910 bases ends in a poly(A) tail. A polyadenylationsignal, ATTAAA, is located 22 nucleotides upstream of thepoly(A) addition site.
Genomic DNA organizationTo analyse the intron-exon organization of NUANCE and toidentify the alternatively spliced isoforms, we searched thegenomic database and found that the NUANCEgene matchedthe publicly available working draft sequences of human cloneswith the accession numbers AL355100.2, AL162832.3,AL359235.2, AL355094.2, AF215937.1, AL352983.2 andAL161756.3 of chromosome 14 in contig NT_025892 mappedto 14q22.1-q22.3 (Fig. 1B). The human NUANCEgene spansover 373 kb. About 6 kb downstream the NUANCEgene is theestrogen receptor 2 gene (ESR2).
The NUANCEgene is split into 115 recognizable exons (Fig.1B, Table 1). All exon-intron boundaries are consistent withthe consensus sequence for splice junctions 5′-GT…AG-3′(Breathnach and Chambon, 1981). The intronic sequencescomprise 94% of the total gene length. The 5′UTR isinterrupted by the longest 55,954 bp intron located 42 bpupstream of the start codon, therefore translation starts in thesecond exon. An alternatively spliced form with the exon 1apositioned 228 bp upstream of exon 1 was found in the ESTBI026470. The exon lengths vary from 50 nt in exon 114 to2101 nt in the exon 48 coding for central coiled-coil sequences.
Exons 3 to 9 encode the ABD region of NUANCE. One ofthe ABD fragments amplified by RACE-PCR (clone 2a)contained an additional exon 8a (Fig. 1C) inserted betweenexons 8 and 9. This leads to premature termination of the ORFand generation of an isoform comprising a truncated ABD,named ABD-S. Although the physiological relevance of ABD-S is not yet clear, the effect of ABD-S overexpression wasanalyzed in transfection experiments (see below; Fig. 9B,E).Several differentially spliced cDNAs corresponding to the3′end of the NUANCE cDNA were found in the non-redundantNCBI database. The cDNAs with accession numbersAK001876, AL080133, AB023228, which correspond to aprotein KIAA1011 (Syne-2), may represent a partial sequenceof NUANCE, since their ORF is not interrupted by any Stopcodons upstream of the methionine proposed as a translationinitiation in AL080133. The cDNA with accession numberAL117404 appears to contain a short isoform with a distinct5′ terminus (Fig. 1D). However, it can also result fromamplification of unprocessed mRNA.
Structural features of NUANCE The NUANCE ORF encodes a protein of 6,885 amino acidswith a molecular mass of 796,000 (Fig. 2). The theoretical pIis 5.29. Three distinct structural domains can be distinguished:the N-terminal ABD (Fig. 3A,B), which is followed by a longlargely helical rod domain (Fig. 3A,E) and a C-terminal TMD(Fig. 3A,D).
The ABD of NUANCE shares a high homology with theABDs of the recently identified proteins enaptin (S. Braune,MD Thesis, University of Cologne, 2001) and calmin (Ishisakiet al., 2001). The ABDs of these three proteins differ from theconventional ones as they have 30 amino-acids long linkersbetween two CH domains (Fig. 3B). Across its 255 amino acidABD region, NUANCE shares about 49% identity withenaptin, 45% with calmin and 43% with β-spectrin, plectin,dystonin and MACF. The phylogenetic analysis of the ABDssuggested that NUANCE, enaptin and calmin form a distinctfamily within the α-actinin superfamily (Fig. 3C). BLASTsearches with human NUANCE cDNA identified severalhighly homologous mouse EST clones. On the basis of thesequence of EST clones AI747790 and AA498987, we havedesigned primers and amplified a partial mouse NUANCEcDNA. The amino-acid sequence of the mouse and humanNUANCE ABD is well conserved with an identity of 97%.
A central rod domain contains coiled coils interrupted byseveral fragments of random coils. Furthermore, four nuclearlocalization signals (amino acids 1188-1205, 1464-1467, 3629-3645 and 6115-6132) and two leucine zippers (amino acids2127-2148 and 5008-5029) were also predicted by computer

3210
analysis within the rod domain (Fig. 3A). Multiple repeatedunits weakly homologous to the triple-helical spectrin-likerepeats of dystrophin and utrophin can be recognized in the
regions predicted for two- and three-stranded coiled coils (Fig.3E). The repeats found in NUANCE are less regular than theones of dystrophin and utrophin (Winder et al., 1995). The
Journal of Cell Science 115 (15)
B
A0 2 4 6 8 10 12 14 16 18 20 kb16
ATG TAA poly A
AA480953
AA761297
1
2
3
4
5
6
7
8
9
10
11
12
13
DKFZp434G173
10 kb
1
ESR2
DKFZp434G173
90 100 115
10 20 30 408a
107
DKFZp434H2235
106a
50 60 70 80
111
1a
11a 2 3 4 5 6 8 a 97 8
0.2 kb
BI026470 NUANCE N-33C
NUANCE
AL117404
AB023228
0.5 kb
106a106 107 108 109 110 111 114 115112 113D
Fig. 1.Cloning of the human NUANCEgene. (A) The 21.8 kb cDNA was assembled from 13 overlapping clones obtained by RT- and RACE-PCR using mRNA from BL-60 cells as the template. (B) The exon-intron organization of the human NUANCEgene. The NUACEgenecontains 114 protein-coding exons. Introns, represented by lines, are drawn to scale; exons are shown as vertical bars. Exon 1a, found in theEST BI026470, exon 8a, found in the short isoform NUANCE-ABD-S, exon 103a, found in the KIAA1011 protein (accession numberAB023228), and the longer form of exon 104, which is found in DKFZp434H2235 mRNA (accession number AL117404), are represented byempty bars. The positions of Start and Stop codons are indicated by arrows. The annotated sequences of DKFZp434G173 (accession numberNM_015180) and DKFZp434H2235 are underlined by solid lines. The estrogen receptor 2 gene (ESR2) located downstream is boxed.Alternative splicing at the 5′ end (C) and at the 3′ end (D) of the NUANCEgene. An additional exon 1a that encodes an alternativetranscriptional start was found in EST BI026470. The boxes represent exons drawn to scale. Gray shading indicate coding regions; white boxesindicate untranslated regions. Aternatively spliced coding exons are shown in black. The locations of the ATG start codon and the TGA stopcodon are indicated. The size of each exon and intron is given in Table 1.

3211An actin-binding protein at the nuclear envelope
alignment of the 22 best-defined NUANCE repeats are shownin Fig. 3E. The helix A is characterized by a highly conservedtryptophan followed by a hydrophobic residue. The first helix(helix A) and the last one (helix C), which are continuous inthe following repeats, are relatively well defined, whereas themiddle helix B is less ordered. Random coils in the regions
4082-4232, 4332-4532 and 6347-6547 (Fig. 3A) may representhinges providing additional flexibility to the molecule.
In addition, the C-terminal region of NUANCE contains a 62amino-acid region similar to the Syne-1 and DrosophilaKlarsicht proteins, which are involved in nuclear migration andnuclear positioning (Apel et al., 2000; Mosley-Bishop et al.,
Exon Sequence at exon-intron boundary Exon Intron no. 5′ splice donor 5′ splice acceptor size (bp) size (bp)
1a AGCCCG CCCCGG/gtaaga 170 2281 GCCAAG CGGACG/gtgggt 161 55,9632 tttcag/TTCACT TGCAAG/gtaatt 130 31,3863 tgatag/CTGAAC GCCAGG/gtaagc 62 1,0184 ctgcag/CACACT CAGTTG/gtaaga 96 945 tcacag/CCTCGG CGATCA/gtaagt 78 886 ttccag/ATTAAG TTTCAT/gtaagt 93 7,6867 ccttag/ATTGAG CGCCAC/gtaagt 182 4,7188 tgatag/CTATGA CAGAAG/gtaaag 197 1,5148a aaccag/CTTACA GACCAG/gtgact 82 5,0129 ttgcag/ATGTGG GCTCAG/gtatgt 101 2,27310 atacag/GGAAAG TATAAT/gtaagt 102 3,70811 ttgcag/AGCCTG CACCAG/gtgact 138 8,61612 tttcag/ATTAAT TTCAAG/gttgga 165 1,11713 tttcag/AGCCTG AAGACG/gtgtgt 113 83414 ttccag/AATCAA TGGCAT/gtaagc 163 1,63915 tcccag/AAATTT ATTTGG/gtaaag 79 24716 ttgtag/CTGGAG AAAGAG/gtattt 189 1,45717 atgtag/GTACCC CAACTT/gtaagt 165 94218 gtatag/GAAATG ATAAAG/gtaaaa 150 2,56919 ttgtag/GCTGGA TTGAAG/gtatgt 162 4,79320 taatag/CATCTT GTCCAG/gtctct 159 37221 ttcaag/ATCAAT CACAAG/gtggga 174 2,71822 ttttag/GAAGCA TGGGAG/gtaaga 135 1,07423 ttgtag/TCTCTT CATGAG/gtacaa 159 1,74424 aattag/GCCTGC AAACGA/gttagt 212 14225 ttctag/GGGGAC GAAAAG/gtgtta 91 82726 ccttag/GCAATG AGAGAG/gtaaac 110 58527 gcccag/GTATGA CTACAG/gtaatt 127 1,52028 atctag/GTCATA TACCAG/gtaaaa 158 1,11429 acccag/AATGGA CTAAGG/gtaagt 148 62630 ttttag/AATATC ACATCG/gtaagt 611 3,70431 atcaag/GAAAAA CGAATG/gtaagg 180 2,74332 tcttag/TCTTGA GCAGAG/gtgagt 151 6,35633 ttacag/ATTGAA CTTGAT/gtaagt 159 96334 tggtag/ATAAAT CTGAAG/gtaatt 162 2,23535 ttttag/GAAGAA TTGCAG/gtaaga 102 1,22136 tttcag/GTCATG CTAGAG/gtgcta 201 34137 atttag/GAGATA TGAATG/gtgagt 172 72238 ctgcag/ATCAAT CTTTCT/gtaaga 113 1,38339 ctgaag/GATTTC ACTGAG/gtagga 203 45240 ttttag/ACTCCA AAAGAG/gtatag 120 9841 ttacag/GGAACA ATCAAG/gtataa 292 1,15742 attcag/GAAATC CCTAAG/gtaaag 152 75343 atttag/CCATCA CTGCAG/gtgagg 310 2,10144 ttttag/GATTCT CTGCAG/gtacta 165 97645 ttgtag/GAACTA AACAAG/gtaagt 342 16,79746 tttcag/GATTCA ATAGAG/gtatgg 156 1,45547 ttttag/AAATTA TAAAGT/gtaagt 266 1,68248 ttctag/AGGACC CGCACA/gtaagt 2,101 2,28649 cactag/AATGTC AGAAAG/gtaatc 323 2,28750 cactag/GTATCT CCAAAG/gtaata 145 5,46651 cttaag/GCCTTG GAAGAA/gtgagt 219 4,99452 aattag/ATTGAA CAACAA/gtaaga 266 3,05853 ttttag/GCTTCT TTTGAG/gtaagt 169 1,65854 ttttag/GAGCTT GAGAAG/gtaata 156 2,50555 tcacag/ATTAGC CAAAAG/gtaaaa 141 1,84956 gaacag/AAAATG AATAAG/gtatgg 183 75457 tcccag/GCCACA GTGCAG/gtaagt 137 6,09658 atctag/ATGGCT GCTGAT/gtaagt 186 1,717
Exon Sequence at exon-intron boundary Exon Intron no. 5′ splice donor 5′ splice acceptor size (bp) size (bp)
59 ttctag/GATGTG GGGGAG/gtaagc 121 1,16860 attaag/GTCATA TTAAAG/gtaagc 183 2,30061 ttatag/GTAGTC CTACAG/gtatgt 132 4,46362 acacag/GGAGAA CGGAAG/gtgggt 198 60063 tttcag/TTGAAC AGCGAT/gtaagg 75 3,11064 tgatag/AATGGA TGTTCA/gtaatg 123 5,43565 ttacag/GAAGCA GAAGAG/gtaagt 117 5,74366 tcttag/GGCACC TGCCAG/gtaaga 231 5,85567 tcctag/GCTATG GATCAG/gtaaaa 183 1,31768 ttgcag/CATCCT CAACAG/gtaatt 135 95969 ttacag/GTTCTG TGCAAG/gtaaaa 122 15270 caaaag/CAACCA CCTCAG/gtcagt 142 2,65171 aaatag/GTTTCC ACACAG/gtagaa 132 1,06672 attcag/GAAAAT TACGAG/gtaggg 153 11873 ctccag/ACGCTG TACCAG/gtatga 210 1,64474 ttacag/AATGAA CTTCAG/gtaatc 102 1,22875 acttag/AAAGCA TTAGAG/gtatgc 120 14676 tttcag/GATGTA CACAAG/gtgaga 201 2,01677 tcatag/GTTTTT TTGCAG/gtattt 174 1,63078 ctctag/AAATGG TACCAG/gtattt 132 3,58679 tattag/CAAATA GCTCAG/gtcagc 197 2,03780 tgttag/TTATAA TTTTTT/gtaagt 133 1,36781 ctttag/GAGTTT CACCAG/gtaagt 183 41882 ttacag/CTTCAA CACAAG/gtaatt 147 1,68383 ttttag/GAGTTT AGAAAG/gtgtgt 177 2,11984 acttag/ATACAA TGTCAG/gtgagg 156 6,63985 ttccag/GATATA ACTCAG/gtacta 153 5,90886 gtctag/GTCAAT CTTCAG/gtaaat 171 45687 tcttag/TCTCTG TAAAAG/gtatgc 131 2,62988 tgacag/GTGGAC ATAAAG/gtgggt 205 1,12589 ctgcag/GAGCTG TTTGAG/gttcat 180 1,70390 ttctag/TTTGTT TTCCTG/gtattg 126 1,82291 aaatag/AATCAT CTGCAG/gttaga 155 10792 ttgtag/TGAGCT TATAAG/gtaaac 145 1,23793 atacag/ATGTTA GAACAG/gtgagc 95 1,25694 ccccag/ACCAGA CTGAAG/gtagtg 235 5,49195 tcttag/AATAAA GTAGAG/gtaaac 195 2,21996 aaatag/ACCTGG ATTAAG/gtattg 126 8,94097 atgcag/GAACTA TTAAGG/gtaagt 156 1,97098 ttgcag/GTGGCC CAGAAG/gtaagg 159 1,42299 tgccag/GACTGC ATCAAG/gtaaga 167 13,563100 gtgtag/GGTGAA CAGCAG/gtggga 163 5,794101 tcacag/GATCTA CATGAA/gtaaga 188 482102 catcag/AATCGA TTTGAG/gtaaac 151 363103 atgcag/GCCTTT CTCAGG/gtgagc 183 1,86104 ttgcag/CATTTC CTGAAT/gtgagg 138 712105 caacag/GGCTTC ACTCCG/gtacgg 195 1,188106 gtttag/GGCTTG TGTCAG/gtaaca 277 1,777106a tttcag/ATGTAG GAAAGC/gtcttc 61107 tttcag/GTAAAT CCCTAT/gtaagt 140 2,010108 ttctag/GGAAAG AGTCAG/gtactg 115 695109 ttccag/GTGCCT CTGCAG/gtgagt 203 1,026110 tggtag/GAGATA TGCCAG/gtacgc 201 938111 tttaag/GACTTC CTAATG/gtaagt 129 1,484111a ttccag/GGCTCT CTAATG/gtaagt 171 1,484112 tttcag/CAACTG ACCCAG/gtgagt 192 1,080113 tttcag/AACCCA GCAAAG/gtaaga 84 448114 acccag/CAGTTC GAGCAG/gtaacg 50 273115 tggcag/GGTCCC GCTTCT 1,114
Table 1. Exon-intron organization of the human NUANCE gene

3212 Journal of Cell Science 115 (15)
1 MASSPELPTE DEQGSWGIDD LHISLQAEQE DT QKKAFTCW I NSQLARHTS PSVISDL FTD I KKGHVLLDL LE VLSGQQLP RDKGSNTFQC RI NIEHALTF 1 01 LRNRSI KLI N IH VTDIIDG N PSIILGLIWT IILH FHIE KL AQTLSCNYNQ PSLDDVSVVD SSPASSPPAK KCSKVQARWQ MSARKALLLW AQEQCATYES 201 VNVTDFKSSW RNGMAFLAII HAL RPDLIDM KSVKHRSNKD NLREAFRIAE QEL KIP RLLE PEDVDVVDPD EKSIMTYVAQ FLQYSKDAPG TGEEAQGKVK 301 DAMGWLTLQK E KLQKLLKDS ENDTYFKKYN SLLS FMESFN EE KKSFLDVL SI KRDLDELD KDHLQLREAW DGLDHQINAW KI KLNYALPP PLHQTEAWLQ 401 EVEELMDEDL SASQDHSQAV TLIQE KMTLF KSLMDRFEHH SNILLT FENK DE NHLPLVPP NKLEEMKRRI NNILE KKFIL LLE FHYYKCL VLGLVDEVKS 501 KLDIWNI KYG SRESVELLLE DWHKFIEE KE FLARLDTSFQ KCGEIYKNLA GECQNI NKQY MMVKSDVCMY RKNI YNVKST LQKVLACWAT YVENLRLLRA 601 CFEETKKEEI KEVPFETLAQ WNLEHATLNE AGNFLVEVSN D VVGSSISKE L RRLNKRWRK L VSKTQLEMN LPLMI KKQDQ PTFDNSGNIL S KEEKATVEF 701 STDMS VELPE NYNQNI KAGE KHEKENEEFT GQLKVAKDVE KLIGQVEIWE AEAKSVLDQD DVDTSMEESL KHLIA KGSMF DELMARSEDM LQMDIQNISS 801 QES FQHVLTT GLQAKIQEAK E KVQI NVVKL IAAL KNLTDV SPDLDIRLKM EESQKELESY MMRAQQLLGQ RESPGELISK H KEALIIS NT KSLAKYLKAV 901 EELKNNVTED I KMSLEEKSR D VCAKWESLH HELSLYVQQL KIDIE KGKLS D NIL KLEKQI NKEKKLI RRG RTKGLI KEHE ACFSEEGCLY QL NHHMEVLR1001 EL CEELPSQK SQQEVKRLLK D YEQKIE RLL KCASEIHMTL QPTAGGTSKN EGTITTSE NR GGDPHSEAPF A KSDNQPSTE KAMEPTMKFS LAS VLRPLQE1101 ESIME KDYSA SI NSLLERYD T YRDILEHHL QNNKFRITSD FSSEEDRSSS CLQAKLTDLQ VI KNETDARW KEFEIISL KL E NHVNDI KKP FVI KERDTLK1201 E RERELQMTL NTRMESLETA LRLVLPVEKA SLLL CGSDLP LHKMAIQGFH LIDAD RI YQH LRNIQDSIA K QIEI CNRLEE PGNFVLKELH PFDLHAMQNI1301 IL KYKTQFEG MNHRVQRSED TLKALEDFLA SL RTAKLSAE PVTDLSASDT QVAQENTLTV KNKEGEIHLM KDKAKHLDKC L KMLDMSFKD AERGDDTSCE1401 NLLDAFSI KL SETHGYGVQE EFTEENKLLE A CI FKNNELL KNIQDVQSQI S KIGL KDPTV PA VKHRKKSL I RLDKVLDEY EEEKRHLQEM ANSLPHFKDG1501 REKTVNQQCQ NTVVLWENTK AL VTECLEQC G RVLELLKQY Q NFKSILTTL IQ KEESVISL QAS YMGKENL KKRIAEIEI V KEEFNEHLEV VDKI NQVCKN1601 LQ FYLNKMKT FEEPPFEKEA NII VDRWLDI NEKTEDYYEN LG RALALWDK L FNLKNVIDE WTEKALQKME LHQLTEEDRE RLKEELQVHE QKTSEFSRRV1701 AEIQFLLQSS EIPLELQ VME SSIL NKMEHV Q KCLTGESNC HALSGSTAEL REDLDQAKTQ IGMTESLLKA LSPSDSLEI F T KLEEIQQQI LQQKHSMILL1801 E NQIGCLTPE LSEL KKQYES VSDLFNTKKS VLQDHFSKLL NDQCKNFNDW FSNI KVNLKE CFESSETKKS VEQKLQKLSD FLTLEGRNSK I KQVDSVLKH1901 VKKHLPKAHV KELISWLVGQ EFELEKMESI CQARAKELED SLQQLLRLQD DHRNLRKWLT NQEEKWKGTE EPGEKTELFC QALARKREQF ES VAQLNNSL2001 KEYGFTEEEE IIMEAT CLMD RYQTLLRQLS EIEEEDKLLP TEDQSFNDLA HDVIHWI KEI KESLMVLNSS EGKMPLEERI Q KI KEIILL K PEGDARIETI2101 MKQAESSEAP L VQKTLTDIS NQWDNTLHLA ST YLSHQEKL LLEGE KYLQS KEDLRLMLIE L KKKQEAGFA LQHGLQEKKA QLKI YKKFLK KAQDLTSLLK2201 EL KSQGNYLL ECTKNPSFSE EPWLEIKHLH ESLLQQLQDS VQNLDGHVRE HDSYQVCVTD L NTTLDNFSK E FVSFSDKPV DQI VVEEKLQ KLQELENRLS2301 LQDGTLKKIL ALA KSVKQNT SS VGQKII KD DI KSLQCKQK DLE NRLASAK QEMECCLNNI L KSKRSTEKK G KFTLPGREK QATSDVQEST QESATVEKLE2401 EDWEI NKDSA VEMAMSKQLS L NAQESMKNT EDERKVNELQ NQPLELDTML RNEQLEEIEK L YTQLEAKKA AI KPLEQTEC L NKTETGALV LH NIG YSAQH2501 LD NLLQALIT L KKNKESQYC VLRDFQEYLA A VESSMKALL TD KESLKVGP LDSVTYLDKI KKFIASIE KE KDSLGNLKI K WENLSNHVTD MDKKLLESQI2601 KQLEHGWEQV EQQIQKKYSQ QVVEYDEFTT LMNKVQDTEI SLQQQQQHLQ LRLKSPEERA GNQSMIALTT DLQATKHGFS VLKGQAELQM KRIWGEKEKK2701 NLEDGINNLK KQWETLEPLH LEAENQI KKC DI RNKMKETI LWA KNLLGEL NPSIPLLPDD ILSQI RKCKV THDGILA RQQ SVESLAEEVK D KVPSLTTYE2801 GGDLNNTLED L RNQYQMLVL KSTQRSQQLE FKLEERSNFF AII RKFQLMV QESETLIIP R VETAATEAEL KHHHVTLEAS QKELQEIDSG ISTHLQELTN2901 I YEELNVFER L FLEDQLKNL KI RTNRIQ RF IQ NTCNEVEH KVKFCRQFHE KTSALQEEAD SIQRNELLLN QEVNKGVKEE I YNLKDRLTA I KCCILQ VLK3001 L KKVFDYIGL NWDFSQLDQL QTQVFEKEKE LEE KI KQLDT FEEEHGKYQA LLSKMRAIDL QI KKMTEVVL KAPDSSPESR RLNAQILSQR IE KAKCLCDE3101 II KKLNENKT FDDSSKEKEI LQI KLNAEEN D KLYKVLQNM VLELSPKELD EKNCQDKLET SLH VLNQI KS QLQQPLLINL EI KHIQNEKD NCGAFQEQVW3201 AEM CSI KAVT AIE KQREENS SEASDVETKL REFEDLQMQL NTSIDL RTNV L NHAYENLTR YKEAVTRAVE SITSLEAIII P YRVDVGNPE ESLEMPLRKQ3301 EELEST VARI QDLTE KLGMI SSPEAKLQLQ YTLQELVSKN SAMKEAFKAQ ETEAERYLEN YKCYRKMEED I YTNLSKMET VLGQSMSSLP LSYREALERL3401 EQS KALVSNL IST KEELMKL RQIL RLLRLR CTENDGICLL KI VSALWEKW LSLLEAAKEW EMWCEELKQE WKFVSEEIER EAIILD NLQE ELPEIS KTKE3501 AATTEELSEL LD CLCQYGEN VEKQQLLLTL LLQRI RSIQN VPESSGAVET VPAFQEITSM KERCNKLLQK VQKNKELVQT EIQE RHSFTK EIIAL KNFFQ3601 QTTTS FQNMA FQDHPEKSEQ FEELQSILKK G KLTFENIME KLRI KYSEMY TI VPAEIESQ VEECRKALED IDE KIS NEVL KSSPSYAMRR KIEEI NNGLH3701 NVEKMLQQKS KNIE KAQEIQ KKMWDELDLW HSKLNELDSE VQDIVEQDPG QAQEWMDNLM IP FQQYQQVS QRAECRTSQL NKATVKMEEY SDLL KSTEAW3801 IE NTSHLLAN PADYDSLRTM SHHASTVQMA LEDSEQKHNL LHSI FMDLED LSII FETDEL TQSIQELSNQ VTALQQKIME SLPQIQ RMAD DVVAIESEVK3901 SME KRVSKI K TILLS KEI FD FSPEEHLKHG EVILE NI RPM KKTIAEI VSY Q VELRLPQTG MKPLPVFQRT NQLLQDIKLL E NVTQEQNEL L KVVI KQTNE4001 WDEEIE NLKQ IL NNYSAQFS LEHMSPDQAD KLPQLQGEIE RMEKQILSL N Q RKEDLLVDL KATVLNLHQH LKQEQEGVER D RLPAVTSEE GGVAERDASE4101 RKLNRRGSMS YLAAVEEEVE ESSVKSDNGD EKAEPSPQSW SSLWKHDKDM EEDRASSSSG TIVQEAYGKI STSD NSMAQI LTPDSLNTEQ GPECSLRPNQ4201 TEEGTTPPIE ADTLDSSDAQ GGLEPRVEKT RPEPTEVLHA CKTQVAELEL WLQQANVAVE PETL NADMQQ VLEQQLVGCQ AMLTEIEHKV A FLLETCKDQ4301 GLGDNGATQH EAEALSLKLK T VKCNLEKVQ MMLQEKHSED QHPTILKKSS EPEHQEALQP VNLSELESI V TE RPQFSRQK D FQQQQVLEL KPMEQKDFI K4401 FVEFNAKKMW PQYCQHDNDT TQESSASNQA SSPENDVPDS ILSPQGQNGD KWQYLHHELS SKI KLPLPQL VEPQVSTNMG ILPS VTMYNF RYPTTEELKT4501 YTTQLEDLRQ EASNLQTQEN MTEEAYI NLD KKLFELFLTL SQ CLSSVEEM LEMPRLYRED GSGQQVHYET LALEL KKLYL ALSD KKGDLL KAMTWPGENT4601 NLLLECFDNL Q VCLEHTQAA AVCRSKSLKA GLDYNRSYQN EI KRLYHQLI KSKTSLQQSL NEISGQSVAE QLQKADAYTV ELE NAESRVA KLRDEGERLH4701 LP YALLQEVY KLGDVLDSMW GMLRARYTEL SSPFVTESQQ DALLQGMVEL VKIG KEKLAH GHLKQTKSKV ALQAQIE NHK VFFQKLVADM LLIQAYSAKI4801 LPSLLQ NRET FWAEQVTEVK ILEE KSRQCG MKLQSLLQKW EEFDENYASL E KDLEILIST LPS VSLVEET EERLVERIS F YQQIKRNIGG KHARLYQTLN4901 EG KQLVASVS CPELEGQIAK LEEQWLSLNK KIDHELHRLQ AL LKHLLSYN RDSDQLTKWL ESSQHTLNYW KEQSLNVSQD LDTI RSNI NN FFEFSKEVDE5001 KSSLKTAVIS IG NQLLHLKE TDTATLRASL AQFEQKWTML ITQLPDIQEK LHQLQMEKLP S RKAITEMIS WMNNVEHQTS DEDSVHSPSS ASQVKHLLQK5101 HKEFRMEMDY KQWIVDFVNQ SLLQLSTCDV ES KRYERTEF AEHLGEMNRQ WHRVHGMLNR KIQHLEQLLE SITESE NKIQ IL NNWMEAQE ERLKTLQKPE5201 S VIS VQKLLL D CQDIENQLA I KSKALDELK QSYLTLESGA VPLLEDTASR IDEL FQKRSS VLTQVNQLKT SMQSVLQEWK I YDQLYDEVN MMTI RFWYCM5301 EHSKPVVLSL ETL RCQVENL QSLQDEAESS EGSWEKLQEV IG KLKGLCPS VAEIIEE KCQ NTHKRWTQVN QAIADQLQKA QSLLQLWKAY S NAHGEAAAR5401 L KQQEAKFQQ LANISMSGNN LAEILPPALQ DI KELQHDVQ KTKEAFLQNS S VLDRLPQPA ESSTHMLLPG PLHSLQRAAY LE KMLLVKAN E FEFVLSQFK5501 D FGVRLESLK GLIMHEEENL D RLHQQEKEN PDSFLNHVLA LTAQSPDIEH L NEVSLKLPL SD VAVKTLQN MNRQWIRATA TALERCSELQ GIGLNEKFLY5601 CCEKWIQLLE KIEEAL KVDV A NSLPELLEQ QKTYKMLEAE VSI NQTIADS YVTQSLQLLD TTEIENRPEF ITE FSKLTDR WQNAVQGVRQ RKGDVDGLVR5701 QWQDFTTSVE NLFRFLTDTS HLLSAVKGQE RFSLYQTRSL IHEL KNKEIH FQRRRTTCAL TLEAGEKLLL TTDL KTKESV G RRISQLQDS WKDMEPQLAE5801 MIKQFQSTVE TWDQCEKKI K EL KSRLQVLK AQSEDPLPEL HEDLHNEKEL I KELEQSLAS WTQNLKELQT MKADLTRHVL VEDVMVLKEQ IEHLH RQWED5901 LCLRVAI RKQ EIED RLNTWV VFNEKNKELC AWLVQMENKV LQTADISIEE MIE KLQKDCM EEI NLFSENK LQL KQMGDQL IKASNKSRAA EIDD KLNKI N6001 DRWQHLFDVI GSRVKKLKET FAFIQQLDKN MSNLRTWLAR IESELS KPVV YDVCDDQEIQ KRLAEQQDLQ RDIEQHSAGV ES VFNI CDVL LHDSDACANE6101 TECDSIQQTT RSLDRRWRNI CAMSMERRMK IE ETWRLWQK FLDDYSRFED WLKSAERTAA CPNSSEVLYT SA KEELKRFE A FQRQIHERL TQLELI NKQY6201 RRLARENRTD TAS RLKQMVH EGNQRWDNLQ RRVTAVLRRL RHFTNQREEF EGTRESIL VW LTEMDLQLTN VEHFSESDAD DKMRQLNGFQ QEITL NTNKI6301 DQLIVFGEQL IQ KSEPLDAV LIEDELEELH RYCQEVFGRV S RFHRRLTSC TPGLEDEKEA SENETDMEDP REIQTDSWRK RGESEEPSSP QSLCHLVAPG6401 HE RSGCETPV S VDSIPLEWD HTGDVGGSSS HEEDEEGPYY SALSGKSISD GHSWHVPDSP SCPDHHYKQM EGDRNVPPVP PASSTPYKPP YGKLLLPPGT6501 DGG KEGPRVL NGNPQQEDGG LAGITEQQSG AFDRWEMIQA QELHNKLKI K Q NLQQLNSDI SAITTWL KKT EAELEMLKMA KPPSDIQEIE L RVKRLQEIL6601 KAFDTYKALV VSVNVSSKEF LQTESPESTE LQSRLRQLSL LWEAAQGAVD SWRGGLRQSL MQCQDFHQLS QNLLLWLASA KNRRQKAHVT DPKADPRALL6701 ECRRELMQLE KELVERQPQV DMLQEISNSL LI KGHGEDCI EAEE KVHVIE KKLKQLREQV SQDLMALQGT QNPASPLPSF DE VDSGDQPP ATSVPAPRAK6801 Q FRAVRTTEG EEETESRVPG STRPQRSFLS RVVRAALPLQ LLLLLLLLLA CLLPSSEEDY S CTQANNFAR S FYPMLRYTN GPPPT
Fig. 2.Deducedpolypeptide sequence ofhuman NUANCE.Amino acid residues arenumbered on the left.The actin bindingdomain of NUANCE ishighlighted in green; 22detected dystrophin-likespectrin repeats are incyan, the transmembraneregion is in yellow,Klarsicht-like domain isin black and underlined.Predicted nuclearlocalization signals areshown in blue and areunderlined, leucinezippers are in red andunderlined.
Fig. 3. Structural features of human NUANCE. (A) The ABD is represented by an empty box; 22 spectrin repeats with considerable homologyto dystrophin are shown as filled ovals; and the TMD is indicated by a black bar. The positions of nuclear localization signals and leucinezippers are indicated. Coiled-coil regions were detected by the MultiCoil program (Wolf et al., 1997) with a window size of 21. Blue and redlines mark the location of predicted dimeric or trimeric coiled coils, respectively. (B) Alignment of the ABDs of NUANCE, enaptin, calmin(BAB59010), β-spectrin (AAA60580) and MACF (AAD32244). NUANCE, enaptin and calmin harbor long stretches between both CHdomains unlike conventional ABDs of β-spectrin and MACF. (C) A phylogenetic tree of the ABDs of NUANCE and other proteins of the α-actinin superfamily on the basis of calculations from ClustalW alignment of these domains. The accession numbers are: human filamin(AF184126), human dystrophin (P11532), Dictyosteliumcortexillin (L49527), human β-spectrin (M96803), Drosophilakakapo (AJ011924),Dictyosteliuminteraptin (AF057019), chicken fimbrin (A37097), mouse α-actinin (P12814), human utrophin (P46939), mouse plectin(AF188012), mouse MACF (AF150755), mouse dystonin (AF252549) and human calmin (BAB59010). (D) Klarsicht-like domain ofNUANCE. The C-termini of NUANCE, human Syne-1 (KIAA0796 protein, BAA34516), mouse Syne-1 (AAG24392), human lymphocytemembrane associated protein (LMAP, AAC02992), an uncharacterised C. elegansprotein similar to myosin-like proteins (AAF40010) and D.melanogasterKlarsicht protein (AAD43129) are aligned with ClustalW version 4.2. The amino acids similar in more than 40% of thesequences are shaded. Three parts of the Klarsicht-like domain are marked. (E) Alignment of 22 selected spectrin repeats of human NUANCE.The multiple alignment was made using the CLUSTAL W program (EMBL). The bars indicate positions of three helices according to thestructure-based alignment of Winder and colleagues (Winder et al., 1995).

3213An actin-binding protein at the nuclear envelope
NUANCESyne -1LM APSyne -1AAAF40010Klarsich t
T E G E E E T E S R V P G S T R P Q R S F L S R V V R A A L P L Q L L L L L L L L L A C L L P S S E E D Y S C T Q A N N F A R S F Y P - - - - - - - M L R Y T N G P P P T - - - - 78T K G G S D S S L S E P G P G R S G R G F L F R V L R A A L P L Q L L L L L L I G L A C L V P M S E E D Y S C A L S N N F A R S F H P - - - - - - - M L R Y T N G P P P L - - - - 78T K G G S D S S L S E P G P G R S G R G F L F R V L R A A L P L Q L L L L L L I G L A C L V P M S E E D Y S C A L S N N F A R S S T P C S D T RM A L L H S E L S R C H L Q K CW 89T R D G S D S S R S D P R P E R V G R A F L F R I L R A A L P F Q L L L L L L I G L T C L V P M S E K D Y S C A L S N N F A R S F H P - - - - - - - M L R Y T N G P P P L - - - - 78S P I P D D S T L S E E Q L R R - Q R S RWR R V L R T A L P L Q A L L V L L M G A A C L V P H C D D E Y C C Q L L N N F A K S F D P - - - - - - - S L E F V N G P P P F - - - - 77L E S S E E D G E G A D P A Q T S K R GWA WR I A R A A V P M Q V A L F T I F C A A C L M Q P N C C D N L N N L S M S F T P Q L R Y I R G P P P I 74
kakapoplectin
MACF
dystonin
dystrophin
utrophin
β-spectrin
α-actinin
interaptin
enaptin
NUANCE
calmin
filamin
fimbrin a fimbrin b
cortexll in
A
B
D
C
E
CH1NUANCE D T Q K K A F T C W I N S Q L A R H T S P S V I S D L F T D I K K G H V L L D L L E V L S G Q Q L P R - D K G S N - - T F Q C R I N I E H A L T F L R - - - - - - - N R S I K L I N 80enaptin I V Q K R T F T K W I N S H L A K R K P P M V V D D L F E D M K D G I K L L A L L E V L S G Q K L P C - E Q G H R V K R I H A V A N I G T A L K F L E G R K S M Y R G S P I K L V N 89calmin N V Q K R T F T R W I N L H L E K C N P P L E V K D L F V D I Q D G K I L M A L L E V L S G R N L L H - E Y K S S S H R I F R L N N I A K A L K F L E - - - - - - - D S N V K L V S 82 -spectrin A V Q K K T F T K W V N S H L A R V S C R - - I T D L Y T D L R D G R M L I K L L E V L S G E R L P K P T K G R M - - R I H C L E N V D K A L Q F L K - - - - - - - E Q R V H L E N 79MACF R V Q K K T F T K W V N K H L M K V R K H - - I N D L Y E D L R D G H N L I S L L E V L S G I K L P R - E K G R M - - R F H R L Q N V Q I A L D F L K - - - - - - - Q R Q V K L V N 78
I H V T D I I D G N P S I I L G L I W T I I L H F H I E K L A Q T L S C N Y N Q P S L D D V S V V - D S S P A S S P P A - - - K K C S K V Q A R W Q M S A R K A L L L W A Q E Q C A 166I N A T D I A D G R P S I V L G L MW T I I L Y F Q I E E L T S N L P Q L Q S L S S S A S S V D S M V S T E T A S P P S - - - - - K R K V A A K I Q G N A K K T L L K W V Q H T A G 174I D A A E I A D G N P S L V L G L I W N I I L F F Q I K E L T G N L S R N - S P S S S L S P G S G G T D S D S S F P P T P T A E R S V A I S V K D Q R K A I K A L L AW V Q R K T - 170M G S H D I V D G N H R L T L G L I W T I I L R F Q I Q - - - - - - - - - - - - - - - - D I S V E T E D N K E - - - - - - - - - - - - - - - - - - K K S A K D A L L L W C Q M K T A 135I R N D D I T D G N P K L T L G L I W T I I L H F Q I S - - - - - - - - - - - - - - - - D I Y I S G E S G D - - - - - - - - - - - - - - - - - - - - M S A K E K L L L W T Q K V T A 132
T Y E S V N V T D F K S S W R N G M A F L A I I H A L R P D L I D M K S V K H R S N K D N L R E A F R I A E Q E L K I P R L L E P E D V D V V D P D E K S I M T Y V A Q F L Q Y S 255K Q M G I E V K D F G K S W R T G L A F H S V I H A I Q P E L V D L E K V K T R S N R E N L E D A F T I A E T Q L G I P R L L D P E D V D V D K P D E K S I M T Y V A Q F L T Q Y 263R K Y G V A V Q D F A G S W R S G L A F L A V I K A I D P S L V D M K Q A L E N S T R E N L E K A F S I A Q D A L H I P R L L E P E D I M V D T P D E Q S I M T Y V A Q F L E R F 259G Y P N V N I H N F T T S W R D G M A F N A L I H K H R P D L I D F D K L K K S N A H Y N L Q N A F N L A E Q H L G L T K L L D P E D I S V D H P D E K S I I T Y V V T Y Y H Y F 224G Y T G V K C T N F S S C W S D G K M F N A L I H R Y R P D L V N M E R V Q V Q S N R E N L E Q A F E V A E R - L G V T R L L D A E D V D V P S P D E K S V I T Y V S S I Y D A F 220
NUANCEenaptincalmin -spectrinMACF
NUANCEenaptincalmin -spectrinMACF
CH1 CH2
CH2
neck region transmembrane region C-terminal tail
0.2
0.4
0.6
0.8
1
0
1000 2000 3000 4000 5000 6000
NLS NLS NLSLZ LZNLS
helix A helix B helix C
rep291_384 - - - - - - T G E E A Q G K V K D A M GW L T L Q K E K L Q - - - - - - - - - K L L K D S E N D Y F K K Y N S L L S F M E S F N E E K K S F L D V L S I K R D L D - - - - - - - - - - - E L D K D H L Q L R E AW D G L D H Q I N AW K I K L 93rep385_475 - - - - - - - N Y A L P P P L H Q T E AW L Q E V E E L M D - - - - E D L S A S Q D H S Q A V T L I Q E K M T L F K S L M D R F E H H S N I L L T F E N K D E N H L P L V P P N - - - - K L E E M K R R I N N I L E 91rep573_662 - - - - - N I Y N V K S T L Q K V L A C W A T Y V E N L R - - - - - - - - - L L R A C F E E T K K E E I K E V P F E T L A QW N L E H A T L N E A G N F L V E V S N D V V G - - - - - S S I S K E L R R L N K R W R K L V 90rep840_934 V S P D L D I R L K M E E S Q K E L E S Y M M R A Q Q L L G Q - - - - - - - - - - - - R E S P G E L I S K H K E A L I I S N T K S L A K Y L K A V E E L K N N V T E D I K M S - - - - - - L E E K S R D V C A K W E S L H H E L S 95rep1616_1711 - - - - - - - - - F E K E A N I I V D R W L D I N E K T E - - - - - - - - - D Y Y E N L G R A L A L W D K L F N L K N V I D E W T E K A L Q K M E L H Q L T E E D R E R L - - - - - - K E E L Q V H E Q K T S E F S R R V A E I Q F L L Q S S E 96rep1942_2037 - - - - L Q Q L L R L Q D D H R N L R K W L T N Q E E K W K - - - - G T E E P G E K T E L F C Q A L A R K R E Q F E S V A Q L N N S L K E Y G F T E E E - - - - - - - - - - - - - - - - E I I M E A T C L M D R Y Q T L L R Q L S E I E E E D K 96rep2038_2130 - - L L P T E D Q S F N D L A H D V I H W I K E I K E S L - - - - - - - - - M V L N S S E G K M P L E E R I Q K I K E I I L L K P E G D A R I E T I M K Q A E S S E A P - - - - - - - - L V Q K T L T D I S N QW D N T L H L A 93rep2211_2304 - - - - - - - - E C T K N P S F S E E P W L E I K H L H E S - - - - - - - - - - - L L Q Q L Q D S V Q N L D G H V R E H D S Y Q V C V T D L N T T L D N F S K E F V S F S D K - - - - P V D Q I V V E E K L Q K L P R L E N R L S L Q D G 94rep2723_2830 - A E N Q I K K C D I R N K M K E T I L W A K N L L G E L N - - - - - - - - - P S I P L L P D D I L S Q I R K C K V T H D G I L A R Q Q S V E S L A E E V K D K V P S L T T - - Y E G G D L N N T L E D L R N Q Y Q M L V L K S T Q R S Q Q L E 108rep4232_4342 - - E P T E V L H A C K T Q V A E L E L W L Q Q A N V A V E P E T L - - - - - - - - - N A D M Q Q V L E Q Q L V G C Q A M L T E I E H K V A F L L E T C K D Q G L G D N G A T Q H E A E A L S L K L K T V K C N L E K V Q M M L Q E K H S E D Q H P 111rep4943_5053 - - - - L K H L L S Y N R D S D Q L T K W L E S S Q H T L N YW K E Q S L N V S Q D L D T I R S N I N N F F E F S K E V D E K S S L K T D V I S I G N Q L L H L K E T D T A T - - - - - - L R A S L A Q F E Q K W T M L I T Q L P D I Q E K L H Q 111rep5054_5177 - - - L Q M E K L P S R K A I T E M I S W M N N V E H Q T S - - D E D S V H S P S S A S Q V K H L L Q K H K E F R M E M D Y K QW I V D F V N Q S L L Q L S T C D V E S K R Y - - E R T E F A E H L G E M N R QW H R V H G M L N R K I Q H L E Q 114rep5282_5381 - - - - I Y D Q L Y D E V N M M T I R F W Y C M E H S K - - - - - - P V V L S L E T L R C Q V E N L Q S L Q D E A E S S E G S W E K L Q E V I G K L K G L C P S V A E I I - - - - - - - - - E E K C Q N T H K R W T Q V N Q A I A D Q L Q K A Q - 101rep5588_5696 - - S E L Q G I G L N E K F L Y C C E K W I Q L L E K - I E - - - - - - E A L K V D V A N S L P E L L E Q Q K T Y K M L E A E V S I N Q T I A D S Y V T Q S L Q L L D T T E I E N R P - E F I T E F S K L T D R W Q N A V Q G V R Q R K G D V D 110rep5697_5808 - - G L V R QW Q D F T T S V E N L F R F L T D T S H L L S - - - - A V K G Q E R F S L Y Q T R S L I H E L K N K E I H F QW R R T T C A L T L E A G E K L L L T T D L K T K - - - - E S V G R R I S Q L Q D S W K D M E P Q L A E M I K Q F Q S T 112rep5809_5909 - V E T W D Q C E K K I K E L K S R L Q V L K A Q S E - - - - - - - D P L P E L H E D L H N E K E L I K E L E Q S L A S W T Q N L K E L Q T M K A D L T R H V L V E D V M V - - - - - - - L K E Q I E H L H R QW E D L C L R V A I R K 101rep5910_6020 - - - - - - - - - - - Q E I E D R L N T W V V F N E K N K - - - - - - - - - - - - - - N K Q Y R R L A R E N R T D T A S R L K Q M V H E G N Q R W D N L S N K S R A A - - - - - - - - - E I D D K L N K I N D R W Q H L F D V I G S R V K K L K E T 88rep6021_6133 - - - - F A F I Q Q L D K N M S N L R T W L A R I E S E L S - - - K P V V Y D V C D D Q E I Q K R L A E Q Q D L Q R D I E Q H S A G V E S V F N I C D V L L H D S D A C A N E - T E C D S I Q Q T T R S L D R R W R N I C A M S M E R R M K I E E 113rep6134_6242 - - - T W R L W Q K F L D D Y S R F E D W L K S A E R T A A - - - - C P N S S E V L Y T S A K E E L K R F E A F Q R Q I H E R L T Q L E L I N K Q Y R R L A R E N R T D T A S - - - - - R L K Q M V H E G N Q R W D N L Q R R V T A V L R R L R H 109rep6243_6347 - - - F T N Q R E E F E G T R E S I L V W L T E M D L Q L T - - - - - - N V E H F S E S D A D D K M R Q L N G F Q Q E I T L N T N K I D Q L I V F G E Q L I Q K S E P L D A V - - - - - L I E D E L E E L H R Y C Q E V F G R V S R F H R R L - - 105rep6547_6658 - L K I K Q N L Q Q L N S D I S A I T T W L K K T E A E L E - - M L K M A K P P S D I Q E I E L R V K R L Q E I L K A F D T Y K A L V V S V N V S S K E F L Q T E S P E S - - - - - - T E L Q S R L R Q L S L L W E A A Q G A V D S W R G G L R Q 112rep6659_6768 - - - S L M Q C Q D F H Q L S Q N L L L W L A S A K N R R Q - - - - - K A H V T D P K A D P R A L L E C R R E L M Q L E - K E L V E R Q P Q V D M L Q E I S N S L L I K G H - G E D C I E A E E K V H V I E K K L K Q L R E Q V S Q D L M A L Q 110
β
β
β
Fig. 3.

3214
1999). The highly conserved hydrophobic stretch of 22 aminoacids (residues 6848 to 6872) in the Klarsicht-like domain wasidentified as a transmembrane region (Fig. 3D). It is flanked bythe N-terminally positioned neck region and the C-terminal tail.
Tissue distribution of NUANCE mRNAIn a Human Tissue Multiple Expression array containingmRNA from a variety of human tissues and cell lines we foundthat most of the tissues showed detectable levels of NUANCEmRNA. The highest expression was detected in the kidney,both adult and fetal, liver, stomach and placenta, and the lowestlevels were in skeletal muscle and brain (Fig. 4A, Table 2). Thecorpus callosum and pituitary gland displayed a relativelystrong signal in contrast to the generally low levels ofexpression in other parts of the brain. Expression of NUANCE
in lymphatic organs was not uniform either. High levels ofmRNA were detected in spleen and lymphatic nodes; howeverthey were relatively low in the thymus and hardly detectablein bone marrow. Taken together with the strong signal seen inperipheral blood lymphocytes, this suggests that NUANCE ischaracteristic for mature lymphocytes. In the digestive systemonly stomach, duodenum and salivary gland showed anelevated level of NUANCE mRNA relative to the other partsof the gastrointestinal tract. In addition, significant amounts ofmRNA were also noted in the trachea, prostate, gonads, thyroidand adrenal glands. In RNA from cancer-derived cell lines,NUANCE was expressed at hardly traceable amounts with theexception of Daudi Burkitt’s lymphoma cells.
NUANCE associates with nucleimAb K20-478 generated against the ABD of NUANCErecognized a protein of about 800 kDa that cofractionated withnuclei and membranes of COS7 cells (Fig. 4B,C). The mAbsagainst lamin B2 as nuclear protein and annexin A7 as mainlycytosolic protein were included as a control. Weaker bandsbelow may represent degradation products of NUANCE,although they could also be short isoforms or crossreactiveproteins. For size estimation, we included the A chain of EHS-laminin with a molecular mass of about 400,000, whichmigrated significantly faster than NUANCE (data not shown).
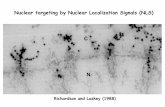
In immunofluorescence studies the mAb yielded a spottedrim-enriched nuclear labeling in Burkitt’s lymphoma cells BL-60, human embryonic kidney cells 293 and COS7 cells (Fig. 5).In addition, a dotted pattern was detected in the cytoplasm,which might result from association with vesicular structures orER (Fig. 5A,C). A similar pattern was observed in C3H/10T1/2mouse fibroblasts (data not shown). NUANCE is also seen
Journal of Cell Science 115 (15)
Fig. 4.Expression analysis of NUANCE. (A) A northern dot-blot ofa variety of human tissues and cell lines, probed with a fragmentcorresponding to the ABD of NUANCE. These results aresummarized in Table 2. Lane 12 represents controls: A12, yeast totalRNA; B12, yeast tRNA; C12, E. coli rRNA; D12, E. coliDNA; E12,poly(A); F12 human C0t_1 DNA; G12, human DNA 100 ng; H12,human DNA 500 ng. (B) Immunoblots of the COS7 cellshomogenate. (C) Cells fractionated into nuclei, cytosol andcytoplasmic membranes. Each fraction was separated on 3-15%gradient SDS-PAGE and immunoblotted with anti-NUANCE, anti-lamin B and anti-annexin A7 mAbs as indicated. The positions ofmolecular mass marker proteins are shown on the left.
Fig. 5. Nuclear localization of NUANCE in COS7 cells (C,G)immunolabeled with the anti-NUANCE mAb K20-478. NUANCEremains associated with NE during prophase (A,B, arrow) and withchromosomes during prometaphase (A,B, large arrowhead). At laterstages NUANCE is diffusely distributed throughout the cytoplasm (A,B, small arrowhead). NUANCE is also detected in bridges, whichwere occasionally seen between the nuclei of two cells (C,D, arrow).DNA was visualized with DAPI (B,D). Bar, 20 µm.

3215An actin-binding protein at the nuclear envelope
associated with bridges that occasionally connected nuclei inCOS7 cells (Fig. 5C,D). The punctate NUANCE staining of thenuclear surface was reminiscent of that of nucleoporins. Acomparison of NUANCE and Nup358 distribution (Wu et al.,1995) however showed that both proteins are only partlycolocalized (Fig. 6A-C), which implies that NUANCE istargeted to the NE without being localized to nuclear pores.
To clarify whether NUANCE is associated with the inner(INM) or outer nuclear membrane (ONM) we comparedthe immunostaining of the digitonin- and Triton X-100-permeabilized cells. Digitonin disrupts the plasmalemmaleaving the intracellular membranes, including the NE, intact(Adam et al., 1990). As a result, the mAb can accessONM-associated antigens but not the proteins facing thenucleoplasm. The anti-NUANCE mAb stained only the
nuclear periphery of the digitonin-permeabilized cells, whichwas similar to the staining for cytoplasm-facing nucleoporin358 (Fig. 6J-L). The anti-lamin B2 mAb taken for a controldid not yield any staining in digitonin-treated cells ininterphasel; instead it labeled mitotic cells on the same coverslip (data not shown). This suggests that NUANCE associateswith the ONM with its N-terminus facing the cytoplasm.However, it does not exclude its further association with theinner membrane of the envelope. In the Triton X-100-treatedcells we also observed a labeling in the nucleus (Fig. 6D-I).It was especially enriched at nucleoli, which were identifiedusing antiserum against the nucleolus protein NO38/B23 (G-I). Furthermore we always observed a diffuse cytoplasmicNUANCE staining that accumulated at the nuclearinvagination harboring the Golgi apparatus (Fig. 6J), as
Expression Position on Tissue type level the grid*
NeurologicalWhole brain tr 1ACerebral cortex tr 1BFrontal lobe tr 1CParietal lobe + 1DOccipital lobe + 1ETemporal lobe + 1FParacentral gyrus tr 1G
of cerebral cortex Pons tr 1HCerebellum, left tr 2ACerebellum, right tr 2BCorpus callosum ++ 2CAmygdala tr 2DCaudate nucleus + 2EHippocampus + 2FMedulla oblongata + 2GPutamen + 2HSubstantia nigra + 3ANucleus accumbens tr 3BThalamus + 3CPituitary gland +++ 3DSpinal cord + 3E
Muscle and heartAorta + 4BHeart + 4AAtrium, left + 4CAtrium, right + 4DVentricle, left + 4EVentricle, right ++ 4FInterventricular septum ++ 4GApex of the heart ++ 4HSkeletal muscle + 7B
Gastro-intestinalEsophagus tr 5AStomach ++++ 5BDuodenum +++ 5CJejunum ++ 5DIleum + 5EIleocecum ++ 5FAppendix ++ 5GColon, ascending + 5HColon, transverse + 6AColon, descending + 6BRectum + 6C
Expression Position on Tissue type level the grid*
Liver ++++ 9APancreas + 9BSalivary gland +++ 9E
Genito-urinarykidney +++++ 7Abladder + 8Cuterus + 8Dprostate ++++ 8Etestis +++ 8Fovary +++ 8G
Lymphoid and hematopoeticSpleen ++++ 7CThymus + 7DLymph node +++ 7FBone morrow + 7GPeripheral blood leukocyte ++++ 7E
PulmonaryTrachea +++ 7HLung ++ 8A
OtherAdrenal gland ++ 9CThyroid gland ++++ 9DMammary gland ++ 9F
Cell linesLeukemia, HL-60 tr 10AHeLa S3 tr 10BLeukemia, K-562 tr 10CLeukemia, MOLT-4 tr 10DBurkitt´s lymphoma, Raji tr 10EBurkitt´s lymphoma, Daudi ++ 10FColorectal adenocarcinoma, + 10G
SW480Lung carcinoma, A549 tr 10H
Fetal and placentaPlacenta ++++ 8BFetal brain tr 11AFetal heart + 11BFetal kidney ++++ 11CFetal liver + 11DFetal spleen + 11EFetal thymus + 11FFetal lung + 11G
Table 2. Expression of NUANCE in 76 human tissues
*Grid references to Fig. 3A.tr, traceable amounts.

3216
visualized with a β-COP-specific mAb (data notshown).
In mitotic cells NUANCE remains associatedwith the NE during its breakdown, which marksthe end of prophase and beginning ofprometaphase (Fig. 5A,B, arrows). Thisindicates a stable association of NUANCE withthe NE. In prometaphase, NUANCE isaccumulated at condensed chromosomes (Fig.5A,B, large arrowheads) and is diffusely presentthroughout the cytoplasm at later stages (Fig.5A, small arrowhead).
Sensitivity to agents affecting thecytoskeletonThe distribution pattern of NUANCE did notoverlap with that of the actin cytoskeleton in spiteof the presence of the predicted conserved ABD(Fig. 7A-C). To test whether disruption of theactin cytoskeleton affects NUANCE localization,we have treated COS7 cells with Latrunculin A(LatA), a drug depolymerizing actin filamentsowing to its G-actin-sequestering activity(Yarmola et al., 2000). Already after 15 minutesexposure to 1 µM LatA, the actin filamentmeshwork was partially disassembled, and actin-rich foci were formed (data not shown). By 30minutes, actin accumulated in the central area ofthe cell in which the NUANCE-positive ‘cloud’was often seen at the invagination of the nuclei(Fig. 7D-F). Along with further disassembly ofthe actin cytoskeleton, the nuclei became lessflattened and acquired an irregular shape withwrinkled invaginations from the side where adiffuse cytoplasmic pool of NUANCE and actinaggregates accumulated (Fig. 7G-I). This effecton nuclei was fully reversible. After 3 hourswashout of LatA their shape was completelyrestored (not shown). This observation impliesthat the nuclear shape is controlled by the actincytoskeleton and that NUANCE may have astructural role in maintaining the nucleararchitecture.
In wound-healing assays performed onconfluent monolayers of COS7 cells, NUANCEwas detected at the leading edge of migratingcells colocalized with F-actin (Fig. 7J-L); thisindicates partial redistribution of NUANCE inpolarized cells.
We have also examined the pattern of NUANCE distributionupon treatment with vincristine and colchicine, drugs thatdisrupt selectively microtubules in the cell. However, nochanges in nuclear morphology or in NUANCE distributionwere observed (data not shown).
Domain analysisThe N-terminal actin-binding domain binds to F-actinin vitro and in vivo
As our immunofluorescence analysis revealed that association
of NUANCE with the actin cytoskeleton is limited to theleading edge in migrating cells and to the perinuclear patchesin LatA-treated cells, we examined the functionality and thesubcellular localization of the ABD. For biochemicalcharacterization of NUANCE we have used the recombinantlyexpressed ABD-containing construct 6×His-ABD. The 6×His-ABD associates with F-actin filaments in a high-speedcosedimentation assay (Fig. 8A). Some 6×His-ABD proteinwas present in the pellet without added actin; however it wasalways enriched in the pellets in the presence of actin-containing samples. The assay was performed with different
Journal of Cell Science 115 (15)
Fig. 6. Localization of NUANCE compared with that of nuclear pores. Cells weredouble labelled with anti-NUANCE (A,D,G,J) and anti-nucleoporin358 (Nup385)(B,E,K) antibodies. A-C, confocal sections of the apical part of a COS7 cell nucleus.The optical surfaces of nuclei are shown with enlargements of the regions indicated.NUANCE- (arrowheads, A-C) and Nup358-positive spots (arrows, A-C) are onlypartially colocalized. (D-L) The equatorial optical sections from the nuclei of COS7cells, treated with 0.5% Triton X-100 (D-I) and 4 µM digitonin (J-L). Visualizationof NUANCE at the NE in digitonin-treated cells (J-L, arrowheads) suggests itslocalization on the cytoplasmic face of the ONM. Note the polar distribution ofcytoplasmic NUANCE (D, J, asterisk) and Nup385. In Triton X-100-treated cells,the intranuclear labeling was enriched in the nucleoli (D-I, arrows), which wasverified by anti-NO38 staining (H). Bar, 5 µm.

3217An actin-binding protein at the nuclear envelope
concentrations of 6×His-ABD in order to quantify binding toF-actin. The Kd value was determined to be 3.8±1 µM, andsaturation was achieved at a 1:1 molar ratio. The presence of6×His-ABD also had an effect on the polymerization kineticsof pyrene-labeled actin (Fig. 8C). It shortened the elongationtime and increased the rate of actin polymerization in aconcentration-dependent manner. Moreover, in low-speed F-actin co-sedimentation assays most of the actin was detectedin the pellet fraction, suggesting that NUANCE ABD acts tobundle F-actin (Fig. 8B).
We expressed GFP-fused ABD in COS7 cells and examinedits subcellular distribution in vivo. In contrast to theendogenous protein, the GFP-ABD containing the 285 N-terminal amino acids of NUANCE was associated with allmicrofilament structures detectable by TRITC-phalloidin,stress fibers, lamellar meshwork and cortical actin (Fig. 9A,D).The GFP-ABD-S fusion protein, which corresponds to a shortalternatively spliced isoform where insertion of an exon 8a
introduced a stop codon in the secondCH domain, showed a slightly differentpattern. The GFP-ABD-S weaklyassociated with the middle parts ofstress fibers but was enriched at theirends, which colocalized with thevinculin-labeled focal adhesions (Fig.9B,E). Interestingly, the small focalcomplexes at the extreme edge oflamellipodia, which were notassociated with stress fibers yet (Nobesand Hall, 1995), did not recruit theGFP-ABD-S (Fig. 9B,E). The GFP-ABD-S association with cortical actinwas mainly confined to the bundles inretracting concave parts but not to theprotruding lamellas. We also noted theformation of multiple spikes andfilopodia in the cells stronglyoverexpressing GFP-ABD or GFP-ABD-S (data not shown). Thefilopodia-rich phenotype waspresumably caused by the increasedformation of actin bundles, which is inagreement with our in vitroobservations. These proteins mighttherefore exhibit a dominant-negativeeffect. Moderate GFP-ABD expressiondid not seem to affect the organizationof the actin cytoskeleton in COS7 cells.
Spectrin repeats1-2 and 15-21 ofNUANCE seem to mediatemembrane targetingTo explore the role of the spectrinrepeats in actin association, weconstructed a fusion protein GFP-ABDsr1-2 harboring a 531 amino acidfragment containing the ABD and thefirst two repeats (Fig. 9C,F). Costainingthe GFP-ABDsr1-2-expressing cellswith TRITC-phalloidin showed overall
colocalization of the fusion protein with filamentous actin (datanot shown). However, the GFP-ABDsr1-2 was stronglyattracted to the subplasmalemmal regions, especially inprotruding lamellas (Fig. 9C,F). This implies a role for the twospectrin repeats in membrane binding. The targeting of thestress fibers and focal contacts was reduced in comparison withthe GFP-ABD and GFP-ABD-S proteins. The anti-NUANCEmAb recognized the GFP-ABDsr1-2 protein (Fig. 9F) as wellas the other two ABD-containing chimeras (data not shown) andshowed an identical pattern.
GFP-sr15-21 harboring spectrin repeats from 15 to 20followed by the random coil stretch and a part of repeat 21were used for the examination of the further spectrin repeats.The distribution pattern of the GFP-sr15-21 suggests itsassociation with the intracellular membranes and withvesicular structures colocalizing with β-COP, a component ofthe COP coat (Fig. 9G,J). No association with the NE wasdetected.
Fig. 7.Effect of LatA treatment on the subcellular distribution of endogenous NUANCE. COS7cells were incubated in the absence (A-C) and presence (D-I) of 1 µM LatA for 30 minutes (D-F) and 60 minutes (G-I). (J-L) COS7 cells were stimulated to migrate by wounding a confluentmonolayer of cells. 6 hours after wounding, the cells were processed for immunofluorescencemicroscopy. Cells were stained with an anti-NUANCE mAb followed by an Alexa-488-conjugated secondary antibody (A,D,G,J) and TRITC-phalloidin (B,E,H,K) to visualize actinfilaments. (C-L) overlays. Bars, 20 µM.

3218
The C-terminal transmembrane domain associates withthe nuclear envelopeThe NUANCE staining pattern appeared to be similar to thesubcellular distribution of Syne-1, a protein that is associatedwith the NE in skeletal, cardiac and smooth muscle cells (Apelet al., 2000). As Syne-1 is highly homologous to the C-terminalpart of NUANCE, the targeting of NUANCE to the NE couldresult from the Klarsicht-like domain at the C-terminus. To testour assumption we have prepared two C-terminal constructs,GFP-Cterm1 and GFP-Cterm2, comprising the Klarsicht-likedomain and the preceding spectrin repeats. The GFP-Cterm1protein possesses spectrin repeats 21 and 22, whereas the GFP-Cterm2 harbors only one complete repeat 22 but contains theadditional 14 amino acids gained from the alternatively splicedexon 111. Both fusion proteins showed clear nuclear rim stainingalong with diffuse cytoplasmic staining in the Golgi area (Fig.9H, shown for GFP-Cterm1). Moreover, both constructs seemedto displace endogenous NUANCE from the NE but did notinterfere with the nucleoplasmic staining (Fig. 9K). In heavilyoverexpressing cells both GFP-Cterm1 and GFP-Cterm2localized in patches at the NE and also showed a reticular orvesicular pattern in the cytoplasm (data not shown). To inspectwhether the overexpression of the GFP-Cterm1 affects thenuclear pore distribution, we stained the transfected cells withthe anti-Nup358 antibody. Confocal analysis reveals a mutuallyexclusive pattern of GFP patches and anti-Nup358 staining (Fig.10A-C), suggesting the displacement of nuclear pores from theregions of GFP-Cterm1 accumulation. The appearance of laminsA/C and B, detected by the mAbs JOL2 and LN43, in contrast,
was not affected in GFP-Cterm1- and GFP-Cterm2-expressingcells (data not shown). The overexpression of the C-terminalconstructs did not hamper mitotic progression in COS7 cells.GFP-Cterm2, like the endogenous protein, remained associatedwith the NE during prophase, whereas the anti-Nup358 stainingwas largely diffuse (Fig. 10D-F), indicating at least partialdisassembly of nuclear pores.
GFP-Cterm2∆tm, which is identical to GFP-Cterm2, butlacks the hydrophobic membrane-spanning sequence and theC-terminal tail, confirmed the importance of the TMD for thenuclear association. This protein was found in all cellularcompartments when overexpressed in COS7 cells. However, incells fixed with methanol, only vesicular staining in thecytoplasm remained (Fig. 9I). No nuclear staining wasdetected, and the localization of the endogenous NUANCE wasnot affected by the overexpression of the GFP-Cterm2∆tm(Fig. 9L). Thus, the TMD is necessary for recruitment to theNE. These results suggest that NUANCE is an integral proteinof the NE that shares the TMD with the other proteinsexhibiting a similar subcellular localization.
Having identified the intracellular locations for N-terminaland C-terminal parts of NUANCE we generated a constructGFP-NUA∆460-6643 where the first 459 amino acids weredirectly fused to the C-terminal 315 amino acids thatcorrespond to the Cterm1 construct. In transfected COS7 cells,the GFP-NUA∆460-6643was arranged in various structures fromfine filaments to large aggregates (Fig. 10G-L). In most of thecells, the GFP-NUA∆460-6643was observed at the NE envelope,enriched at one side of the nucleus (Fig. 10J-L). Actin wasoften attracted to the GFP-NUA∆460-6643 fibers and aggregates;however, GFP-NUA∆460-6643 was never seen in associationwith the dynamic submembranous actin cytoskeleton.
DiscussionNUANCE, a transmembrane protein associated withactin and the nuclear envelopeCloning of the NUANCE cDNA yielded the largest protein of
Journal of Cell Science 115 (15)
Fig. 9. Localization of GFP-fused chimeric proteins in transfectedCOS7 cells. Protein domains are presented as in Fig. 3. All GFP fusionprotein comprising ABDs (A-F) were colocalized with actin stressfibers (A,C,D,F, big arrows) and actin meshwork throughout the cell aswell as with the cortical actin in lamellae (C,F, arrowheads). The GFP-ABD-S was enriched at the tips of stress fibers (B,arrow) anddiminished at the plasmalemma (B), whereas the GFP-ABDsr1-2 (C)associated stronger with the submembrane actin. Nascent focalcomplexes did not attract GFP-ABD-S (small arrowheads). The aminoacids AYKN at the C-terminus of the GFP-ABD-S protein wereencoded by the additional exon 8a found by PCR analysis. GFP-ABDsr1-2 protein (C,F) harbors two spectrin repeats in addition to theABD. GFP-sr15-21, corresponding to the middle part of NUANCEfrom spectrin repeat 15 to 20, seems to target intracellular membranesand vesicular structures colocalizing with β-COP-positive structures(G,J, arrow) but not to the NE. By contrast, the GFP-Cterm1 proteinharboring the TMD together with the two preceding spectrin repeats isrecruited to the NE and adjacent ER (H). Note the lack of nuclear rimstaining in the transfected cell (K, arrowhead). The GFP-Cterm2∆tmfusion protein is associated with vesicles but not with the NE (I,L,arrowheads). Cells transfected with the GFP-Cterm2∆tm constructwere fixed with methanol (see details in the text). The transfectedCOS7 cells were double-labeled with TRITC-phalloidin (D), vinculin(E) and NUANCE (F,K,L) and β-COP (J) mAbs. Bar, 20 µm.
Fig. 8. Binding of 6×His-ABD to actin in vitro. (A) Cosedimentationof 10 µM 6×His-ABD with 2.5 µM F-actin by ultracentrifugation. (B)Bundling of 10 µM actin in the presence of 6×His-ABD analyzed bylow-speed centrifugation. S, supernatant; P, pellet. Positions of ABDand actin are indicated on the right. (C) The effect on polymerizationkinetics of 8 µM actin in the absence or presence of various amountsof 6×His-ABD observed by the change in pyrenyl-actin fluorescence.

3219An actin-binding protein at the nuclear envelope
the α-actinin superfamily that has so far been characterized. Itis conceivable that NUANCE forms dimers or even higherorder oligomers since it has an extremely long largely coiled-coil rod with two leucine-zippers. NUANCE is localized at the
NE, which is rather unusual for actin-binding proteins. TheNUANCE ABD appears to bind to F-actin in vitro and in vivoand promote actin bundling. Moreover, transfection with theGFP-NUA∆460-6643 mutant has resulted in significant

3220
recruitment of F-actin to the nuclearmembrane. This difference between theNUA∆460-6643and wild-type phenotypesindicates that the ABD of the full-lengthNUANCE could be regulated in vivo, forinstance by adopting a conformationwith the actin-binding sites beinghidden. Two other proteins, calmin andDictyosteliuminteraptin, which likewisepossess ABDs at the N-termini andTMDs at the C-termini, associate withcytoplasmic reticular structures and, inthe case of interaptin, with the NE(Rivero et al. 1998; Ishisaki et al., 2001).The predicted TMDs of these threeproteins show very low homology andmay be responsible for the differentialintracellular localization.
NUANCE appears to be recruited tothe NE through the Klarsicht-like C-terminal domain harboring a TMD.DrosophilaKlarsicht protein is requiredfor migration of nuclei in developingretinal photoreceptors (Mosley-Bishopet al., 1999). Similarly, a role in themigration of micronuclei in myotubesand/or their anchoring at thepostsynaptic apparatus was ascribed toSyne-1 (Apel et al., 2000), a protein thatis highly homologous to the C-terminalpart of NUANCE. Syne-1 is selectivelyassociated with the nuclei that liebeneath the postsynaptic membrane atthe neuromuscular junctions in skeletal,cardiac and smooth muscle cells. Syne-1is strongly expressed in the muscle andbrain, tissues with the lowest level ofNUANCE expression. Since the N-terminus of the longest Syne-1 isoformhas not yet been identified, it is plausiblethat the full-length Syne-1 protein alsopossesses an N-terminal ABD andtherefore may represent a muscle/braincounterpart of NUANCE. Nuclearpositioning and migration are essentialfor the movement of pronuclei duringfertilization, normal mitotic and meioticcell division and various morphogeneticprocesses during metazoan development.Nuclear migration has been directlylinked to a human disease, the braindevelopmental disorder lissencephaly(Morris, 2000). Migration of neuronsinvolves nucleokinesis, a processwhereby cells acquire an elongatedshape or extend a frontal protrusion, andthe nucleus migrates to its new positionwithin the cytoplasm. The nuclear translocation within the cellcan also be associated with cell motility during tumor cellinvasion. In moving fibroblasts, the classical model for cellmotility studies, the nucleus remains centrally anchored within
the cell despite drastic morphological changes. However, thehuman lung adenocarcinoma cells, which generally display afibroblast-like motility, can be switched to the characteristicpattern of cell translocation with nucleokinesis as a distinct
Journal of Cell Science 115 (15)
Fig. 10.The C-terminal domain of NUANCE targets the NE. (A-C) Confocal optical sectionthrough the apical part of the COS7 nucleus showing that the overexpressed GFP-Cterm1protein was arranged in patches displacing nuclear pores (arrows), detected by anti-Nup358antibody (B). (D-F) The GFP-Cterm2 remained associated with the NE in prophase (D,arrow) when the nuclear pores begin to disassemble as indicated by diffuse anti-Nup358staining (E). DNA was visualized with DAPI (F,I,L). (J-L) Two examples of the cellularlocalization of the GFP-NUA∆460-6643overexpressed in COS7 cells stained with TRITC-phalloidin (H,K). The GFP-NUA∆460-6643possesses the N-terminal ABD and the C-terminalKlarsicht-like domain but lacks most of the central rod domain. The GFP-NUA∆460-6643isarranged in fibers (G-I, small arrowheads) or perinuclear aggregates (J-L, arrows) partiallycolocalizing with actin. Dynamic cortical cytoskeleton (G-I, arrow) and some of the stressfibers (G-I, large arrowhead) do not recruit GFP-NUA∆460-6643. (I,L) show overlays of theGFP, TRITC and DAPI channels. Bars (A-C) 5 µM; (D-F) 10 µM; (J-L) 20 µM.

3221An actin-binding protein at the nuclear envelope
step in response to an autocrine motility factor (Klominek etal., 1991). The motility steps of the invasion process have beenproposed to comprise protrusion of the tumor cell pseudopodiathrough surrounding tissues, followed by nucleokinesis and,finally, retraction of the pseudopodia behind the nucleus.
On the basis of the data accumulated on NUANCE, we canenvision several roles for it. Rapid shrinkage of the nucleiaccompanied by the accumulation of cytoplasmic NUANCEand actin at the side of the nucleus in the LatA-treated cellssuggests that the actin cytoskeleton is implicated in controllingnuclear shape and that this process could be mediated byNUANCE. In addition to a role in nuclear positioning and/ormigration, NUANCE may also contribute to the mechanicalconnection between cell surface receptors, cytoskeletalfilaments and nuclear scaffolds. Such a coordinated responseto mechanical stress was demonstrated by micromanipulationof integrins, which resulted in changes in nuclear shape thatmay influence gene expression (Chicurel et al., 1998).Maintenance of the nuclear shape and polarity could beessential for many cell types undergoing mechanical stress invivo, as for instance, for lymphocytes when transgressing theblood vessels.
Eight integral proteins of the NE have so far been identified.Six of them are located at the INM. These are lamina-associated polypepetide (LAP)1 (Senior and Gerace, 1988),LAP2/thymopoietin (Foisner and Gerace, 1993), p58/lamin Breceptor (LBR) (Worman et al., 1988), emerin (Nagano et al.,1996), nurim (Rolls et al., 1999) and MAN1 (Lin et al., 2000).Two integral proteins, POM121and gp210, have been found tobe specific for the nuclear pore membrane (Gerace et al., 1982;Soderqvist and Hallberg, 1994). All of them are arranged withtheir N-termini facing the nucleoplasm. The discovery ofNUANCE as the first example of a protein, which seems toassociate specifically with the ONM, implies that the ONMrepresents a membrane subdomain that is biochemically andfunctionally distinct from the peripheral ER. Recent studiesdemonstrated that the dynein-dynactin complex is recruited tothe cytoplasmic face of the NE during late G2 and earlyprophase prior to the NE breakdown and plays a role in tearingthe NE at the beginning of mitosis (Beaudouin et al., 2002;Salina et al., 2002). Our data suggest that the N-terminal partof NUANCE faces the cytoplasm thereby allowing the proteinto represent a platform for anchoring the dynein-dynactincomplexes at the ONM. Interestingly, the NE wrinklesobserved at the sites of invaginations of the ‘kidney bean’-shaped nuclei in the LatA-treated cells resemble themicrotubules containing finger-like projections of the prophaseand prometaphase cells. Furthermore, accumulation of F-actinand the cytoplasmic NUANCE at the sites of nuclearinvaginations upon LatA treatment imply a link betweenNUANCE and pericentrosomal astral complex. Although atpresent we have no evidence of a functional relationshipbetween NUANCE and NE breakdown, it is possible thatNUANCE is involved in the spatial organization ofmicrotubule-dependent machinery linking it to the NE.
Our results with digitonin-treated cells do not exclude anassociation of NUANCE with the INM where it might face thenucleoplasm with its ABD and interact with nuclear actinwhose role is still elusive (Rando et al., 2000). NUANCEstaining was also observed throughout the nucleoplasm and innucleoli. These forms might well represent differentially
spliced variants lacking the TMD. Such isoforms may build afibrous meshwork in the nucleoplasm by oligomerizating vialeucine zippers of the coiled coil. Although we have notidentified TMD-lacking transcripts in our screen yet, theirexistence is quite plausible since many membrane proteinsincluding calmin are differentially spliced and give rise tosoluble isoforms (Ishisaki et al., 2001). It can be speculatedthat NUANCE plays a role in the organization of theintranuclear space. It was shown that actin is associated withthe nucleoplasmic filaments of nuclear pore complexes and isinvolved in nuclear export (Hofmann et al., 2001). A role ofNUANCE in nuclear export would explain its partialcolocalization with nuclear pores. Actin may also be importantfor packaging the RNA into a Balbiani ring particles and theirtranslocation through the nuclear pores to the cytoplasm(Percipalle et al., 2001). In a complex with actin, NUANCEmay play a role in location and transportation of proteins andRNA within the nucleoplasm. The observation of NUANCE innucleoli is particularly intriguing. The nucleoli are structuresresponsible mainly for ribosomal biogenesis; however theyalso participate in processing or export of some mRNAs andtRNAs (Schneiter et al., 1995; Bertrand et al., 1998) and areinvolved in sequestering cell cycle regulating proteins (Visintinand Amon, 2000). NUANCE would make an ideal candidatefor organizing the nucleoli matrix and ensuring itscompartmentalization and integrity.
This work was supported by a grant from the Center for MolecularMedicine Cologne and by a Köln Fortune studentship to T.L. Wethank Elias Coutavas and Günter Blobel (The Rockefeller University,USA) for providing anti-Nup358 antibodies, Frans Ramaekers and JosBroers (University of Maastricht, the Netherlands) for the gift of anti-lamin A/C and anti-lamin B1 antibodies, Marion Schmidt-Zachmannfor the gift of anti-NO38/B23 antiserum, Mats Paulsson (Universityof Cologne) for laminin, Michael Schleicher (University of Munich,Germany) for sharing reagents and Prof. Diehl (University ofCologne, Germany) for providing the lymphoma cell lines.
ReferencesAdam, S. A., Marr, R. S. and Gerace, L.(1990). Nuclear protein import in
permeabilized mammalian cells requires soluble cytoplasmic factors. J. CellBiol. 111, 807-816.
Ahn, A. H. and Kunkel, L. M. (1993). The structural and functional diversityof dystrophin. Nat. Genet. 3, 283-291.
Apel, E. D., Lewis, R. M., Grady, R. M. and Sanes, J. R.(2000). Syne-1, adystrophin- and Klarsicht-related protein associated with synaptic nuclei atthe neuromuscular junction. J. Biol. Chem. 275, 31986-31995.
Beaudouin, J., Gerlich, D., Daigle, N., Eils, R. and Ellenberg, J.(2002).Nuclear envelope breakdown proceeds by microtubule-induced tearing ofthe lamina. Cell 108, 83-96.
Bertrand, E., Houser-Scott, F., Kendall, A., Singer, R. H. and Engelke, D.R. (1998). Nucleolar localization of early tRNA processing. Genes Dev. 12,2463-2468.
Breathnach, R. and Chambon, P.(1981). Organization and expression ofeucaryotic split genes coding for proteins. Annu. Rev. Biochem. 50, 349-383.
Brown, A., Bernier, G., Mathieu, M., Rossant, J. and Kothary, R.(1995).The mouse dystonia musculorum gene is a neural isoform of bullouspemphigoid antigen 1. Nat. Genet. 10, 301-306.
Chicurel, M. E., Singer, R. H., Meyer, C. J. and Ingber, D. E.(1998).Integrin binding and mechanical tension induce movement of mRNA andribosomes to focal adhesions. Nature392, 730-733.
Foisner, R. and Gerace, L.(1993). Integral membrane proteins of the nuclearenvelope interact with lamins and chromosomes, and binding is modulatedby mitotic phosphorylation. Cell 73, 1267-1279.
Fuchs, E. and Yang, Y.(1999). Crossroads on cytoskeletal highways. Cell 98,547-550.

3222
Gerace, L., Ottaviano, Y. and Kondor-Koch, C.(1982). Identification of amajor polypeptide of the nuclear pore complex. J. Cell Biol. 95, 826-837.
Gregory, S. L. and Brown, N. H.(1998). kakapo, a gene required for adhesionbetween and within cell layers in Drosophila, encodes a large cytoskeletallinker protein related to plectin and dystrophin. J. Cell Biol. 143, 1271-1282.
Hofmann, W., Reichart, B., Ewald, A., Muller, E., Schmitt, I., Stauber, R.H., Lottspeich, F., Jockusch, B. M., Scheer, U., Hauber, J. andDabauvalle, M. C. (2001). Cofactor requirements for nuclear export of Revresponse element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152,895-910.
Ishisaki, Z., Takaishi, M., Furuta, I. and Huh, N. (2001). Calmin, a proteinwith calponin homology and transmembrane domains expressed in maturingspermatogenic cells. Genomics74, 172-179.
Karakesisoglou, I., Yang, Y. and Fuchs, E.(2000). An epidermal plakin thatintegrates actin and microtubule networks at cellular junctions. J. Cell Biol.149, 195-208.
Klominek, J., Sundqvist, K. G. and Robert, K. H.(1991). Nucleokinesis:distinct pattern of cell translocation in response to an autocrine motilityfactor-like substance or fibronectin. Proc. Natl. Acad. Sci. USA88, 3902-3906.
Korenbaum, E., Nordberg, P., Bjorkegren-Sjogren, C., Schutt, C. E.,Lindberg, U. and Karlsson, R. (1998). The role of profilin in actinpolymerization and nucleotide exchange. Biochemistry37, 9274-9283.
Kozak, M. (1987). An analysis of 5′-noncoding sequences from 699 vertebratemessenger RNAs. Nucleic Acids Res. 15, 8125-8148.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assemblyof the head of bacteriophage T4. Nature227, 680-685.
Leung, C. L., Sun, D., Zheng, M., Knowles, D. R. and Liem, R. K.(1999).Microtubule actin cross-linking factor (MACF): a hybrid of dystonin anddystrophin that can interact with the actin and microtubule cytoskeletons. J.Cell Biol. 147, 1275-1286.
Leung, C. L., Liem, R. K., Parry, D. A. and Green, K. J.(2001a). The plakinfamily. J. Cell Sci. 114, 3409-3410.
Leung, C. L., Zheng, M., Prater, S. M. and Liem, R. K.(2001b). TheBPAG1 locus: Alternative splicing produces multiple isoforms with distinctcytoskeletal linker domains, including predominant isoforms in neurons andmuscles. J. Cell Biol. 154, 691-697.
Lin, F., Blake, D. L., Callebaut, I., Skerjanc, I. S., Holmer, L., McBurney,M. W., Paulin-Levasseur, M. and Worman, H. J.(2000). MAN1, an innernuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275, 4840-4847.
Matsudaira, P. (1994). Actin crosslinking proteins at the leading edge. Semin.Cell Biol. 5, 165-174.
Morris, N. R. (2000). Nuclear migration. From fungi to the mammalian brain.J. Cell Biol. 148, 1097-1101.
Mosley-Bishop, K. L., Li, Q., Patterson, L. and Fischer, J. A.(1999).Molecular analysis of the klarsicht gene and its role in nuclear migrationwithin differentiating cells of the Drosophila eye. Curr. Biol. 9, 1211-1220.
Nagano, A., Koga, R., Ogawa, M., Kurano, Y., Kawada, J., Okada, R.,Hayashi, Y. K., Tsukahara, T. and Arahata, K.(1996). Emerin deficiencyat the nuclear membrane in patients with Emery-Dreifuss musculardystrophy. Nat. Genet. 12, 254-259.
Nobes, C. D. and Hall, A.(1995). Rho, rac and cdc42 GTPases regulate theassembly of multimolecular focal complexes associated with actin stressfibres, lamellipodia and filopodia. Cell 81, 53-62.
Okuda, T., Matsuda, S., Nakatsugawa, S., Ichigotani, Y., Iwahashi, N.,Takahashi, M., Ishigaki, T. and Hamaguchi, M. (1999). Molecularcloning of macrophin, a human homologue of Drosophila kakapo with aclose structural similarity to plectin and dystrophin. Biochem. Biophys. Res.Commun. 264, 568-574.
Olski, T. M., Noegel, A. A. and Korenbaum, E.(2001). Parvin, a 42 kDafocal adhesion protein, related to the alpha-actinin superfamily. J. Cell Sci.114, 525-538.
Pascual, J., Castresana, J. and Saraste, M.(1997). Evolution of the spectrinrepeat. Bioessays19, 811-817.
Pepperkok, R., Scheel, J., Horstmann, H., Hauri, H. P., Griffiths, G. andKreis, T. E. (1993). Beta-COP is essential for biosynthetic membranetransport from the endoplasmic reticulum to the Golgi complex in vivo. Cell74, 71-82.
Percipalle, P., Zhao, J., Pope, B., Weeds, A., Lindberg, U. and Daneholt,
B. (2001). Actin bound to the heterogeneous nuclear ribonucleoproteinhrp36 is associated with Balbiani ring mRNA from the gene to polysomes.J. Cell Biol. 153, 229-236.
Prokop, A., Uhler, J., Roote, J. and Bate, M.(1998). The kakapo mutationaffects terminal arborization and central dendritic sprouting of Drosophilamotorneurons. J. Cell Biol. 143, 1283-1294.
Puius, Y. A., Mahoney, N. M. and Almo, S. C.(1998). The modular structureof actin-regulatory proteins. Curr. Opin. Cell Biol. 10, 23-34.
Rando, O. J., Zhao, K. and Crabtree, G. R.(2000). Searching for a functionfor nuclear actin. Trends Cell Biol. 10, 92-97.
Rivero, F., Kuspa, A., Brokamp, R., Matzner, M. and Noegel, A. A. (1998).Interaptin, an actin-binding protein of the alpha-actinin superfamily inDictyostelium discoideum, is developmentally and cAMP-regulated andassociates with intracellular membane compartments. J. Cell Biol. 142, 735-750.
Rolls, M. M., Stein, P. A., Taylor, S. S., Ha, E., McKeon, F. and Rapoport,T. A. (1999). A visual screen of a GFP-fusion library identifies a new typeof nuclear envelope membrane protein. J. Cell Biol. 146, 29-44.
Ruhrberg, C. and Watt, F. M. (1997). The plakin family: versatile organizersof cytoskeletal architecture. Curr. Opin. Genet. Dev. 7, 392-397.
Salina, D., Bodoor, K., Eckley, D. M., Schroer, T. A., Rattner, J. B. andBurke, B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelopebreakdown. Cell 108, 97-107.
Schneiter, R., Kadowaki, T. and Tartakoff, A. M. (1995). mRNA transportin yeast: time to reinvestigate the functions of the nucleolus. Mol. Biol. Cell6, 357-370.
Selbert, S., Fischer, P., Pongratz, D., Stewart, M. and Noegel, A. A.(1995).Expression and localization of annexin VII (synexin) in muscle cells. J. CellSci. 108, 85-95.
Senior, A. and Gerace, L.(1988). Integral membrane proteins specific to theinner nuclear membrane and associated with the nuclear lamina. J. Cell Biol.107, 2029-2036.
Soderqvist, H. and Hallberg, E.(1994). The large C-terminal region of theintegral pore membrane protein, POM121, is facing the nuclear porecomplex. Eur. J. Cell Biol. 64, 186-191.
Strumpf, D. and Volk, T. (1998). Kakapo, a novel cytoskeletal-associatedprotein is essential for the restricted localization of the neuregulin-likefactor, vein, at the muscle-tendon junction site. J. Cell Biol. 143, 1259-1270.
Sun, Y., Zhang, J., Kraeft, S. K., Auclair, D., Chang, M. S., Liu, Y.,Sutherland, R., Salgia, R., Griffin, J. D., Ferland, L. H. et al. (1999).Molecular cloning and characterization of human trabeculin-alpha, a giantprotein defining a new family of actin-binding proteins. J. Biol. Chem. 274,33522-33530.
Tinsley, J. M., Blake, D. J., Roche, A., Fairbrother, U., Riss, J., Byth, B.C., Knight, A. E., Kendrick-Jones, J., Suthers, G. K., Love, D. R. et al.(1992). Primary structure of dystrophin-related protein. Nature 360, 591-593.
Visintin, R. and Amon, A. (2000). The nucleolus: the magician’s hat for cellcycle tricks. Curr. Opin. Cell Biol. 12, 372-377.
Wiche, G. (1998). Domain structure and transcript diversity of plectin. Biol.Bull. 194, 381-383.
Wiche, G., Becker, B., Luber, K., Weitzer, G., Castanon, M. J.,Hauptmann, R., Stratowa, C. and Stewart, M.(1991). Cloning andsequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J. Cell Biol.114, 83-99.
Winder, S. J., Gibson, T. J. and Kendrick-Jones, J.(1995). Dystrophin andutrophin: the missing links! FEBS Lett. 369, 27-33.
Wolf, E., Kim, P. S. and Berger, B. (1997). MultiCoil: a program forpredicting two- and three-stranded coiled coils. Protein Sci. 6, 1179-1189.
Worman, H. J., Yuan, J., Blobel, G. and Georgatos, S. D.(1988). A laminB receptor in the nuclear envelope. Proc. Natl. Acad. Sci. USA85, 8531-8534.
Wu, J., Matunis, M. J., Kraemer, D., Blobel, G. and Coutavas, E.(1995).Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and aleucine-rich region. J. Biol. Chem. 270, 14209-14213.
Yarmola, E. G., Somasundaram, T., Boring, T. A., Spector, I. and Bubb,M. R. (2000). Actin-latrunculin A structure and function. Differentialmodulation of actin-binding protein function by latrunculin A. J. Biol.Chem. 275, 28120-28127.
Journal of Cell Science 115 (15)