Nitrogenase enzyme complexnature.berkeley.edu/brunslab/espm131/powerpoints/N-fix2.pdf ·...
Transcript of Nitrogenase enzyme complexnature.berkeley.edu/brunslab/espm131/powerpoints/N-fix2.pdf ·...
-
Redrawn from www.asahi-net.or.jp/~it6i-wtnb/BNF.html
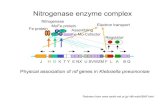
Nitrogenase enzyme complex
Physical association of nif genes in Klebsiella pneumoniae
NitrogenaseMoFe protein
Fe proteinElectron transport
AssemblingFe-Mo-Cofactor
Regulator
H D K T Y E NX U SVWZM F L A B QJ
-
Redrawn from http://www.science.siu.edu/microbiology/micr425/425Notes/12-NitrFix.html
NtrC NtrC
in absence ofN-compounds
NtrB
ADP ATP
nifLA operonnitrC binding site 54 = nitrAbinding site
nif structural genes 54 = nitrAbinding site
nifA binding site
P
Regulation of nitrogen fixation (K. pneumoniae)
-
Nitrogen present, no transcription
Function of NtrA, 54 , thenitrogen factor
-
Nitrogen absent, NtrB phosphorylates NtrC,which activates RNA polymerase
Function of NtrA, 54 , thenitrogen factor
P
-
Redrawn from http://www.science.siu.edu/microbiology/micr425/425Notes/12-NitrFix.html
NtrC NtrC
in absence ofN-compounds
NtrB
ADP ATP
nifLA operonnitrC binding site 54 = nitrAbinding site
nif structural genes 54 = nitrAbinding site
nifA binding site
P
Regulation of nitrogen fixation (K. pneumoniae)
-
Redrawn from http://www.science.siu.edu/microbiology/micr425/425Notes/12-NitrFix.html
NifANifANifL
transcription
NtrC
in absence ofN-compounds
NtrB
ADP ATP
nifLA operon54 = nitrAbinding site
NtrC P
N-compound regulation of NifLA operon
nif structural genes 54 = nitrAbinding site
nifA binding site
-
Redrawn from http://www.science.siu.edu/microbiology/micr425/425Notes/12-NitrFix.html
NifANifA
NifL
transcription
transcription
NtrC
in absence ofN-compounds
NtrB
ADP ATP
nifLA operon54 = nitrAbinding site
nif structural genes 54 = nitrAbinding site
N-compound regulation of NifLA operon
NtrC P
-
Redrawn from http://www.science.siu.edu/microbiology/micr425/425Notes/12-NitrFix.html
NifANifANifANifA
NifLNifLin presence
of O2 orN-compounds
transcription
NtrC
in absence ofN-compounds
NtrB
ADP ATP
nifLA operon54 = nitrAbinding site
nif structural genes 54 = nitrAbinding site
nifA binding site
Oxygen and N-compound regulation of nif structural genes via nifL
NtrC P
-
Measuring N2 fixation rates
-
Acetylene reduction assay
Football has been filled withacetylene
Glass jars contain the plantsamples being evaluated
Sterile vacutainers (normallyused to collect blood) areused to take the gas samplefollowing incubation
Several hundred samplescan be taken each day
http://www.soils.umn.edu/academics/classes/soil3612/Nitrogen_Fixation/Measurement.htm
-
Acetylene reduction assay
A typical trace followinggas chromatography
The greatest peaks areof residual acetylene
Those next to them theethylene peak
http://www.soils.umn.edu/academics/classes/soil3612/Nitrogen_Fixation/Measurement.htm
-
Hydrogen evolution assay
Reduction of dinitrogen to ammonia
N2 + 8H+ + 8e- 2NH3 + H2
H2 is evolved at ratio of 1 moleculeper 2 molecules of N2 reduced
So, can use hydrogen sensor tomeasure H2 evolution to quantify N2fixation
-
Hydrogen evolution assay
Reduction of dinitrogen to ammonia
N2 + 8H+ + 8e- 2NH3 + H2
H2 is evolved at ratio of 1 moleculeper 2 molecules of N2 reduced
So, can use hydrogen sensor tomeasure H2 evolution to quantify N2fixation
-
Hydrogen evolution assay
-
The operation of nitrogenase. The iron protein (Fe) takes electronsfrom central metabolism electron carriers and transfers them to themolybdenum iron protein (MoFe) expending a fair amount of ATP.N2 is converted to ammonia and the electrons in H2 are recycled byhydrogenase. (D. Benson)
-
Stable isotope assays
GC Mass separationCombustion
detector
ion source
magnet
-
mass 29 (15N14N)
ion source
magnet
mass 30 (15N2)
mass 28(14N2)
Mass Separation
-
Stable isotope lab
-
Crop Common name 15N
Agropyrontrichophorum
Pubescentwheatgrass
5.13
A.elongatum Tall wheatgrass 3.04
A.dasystachyum NorthernWheatgrass
3.00
Elymus angustus Wild ryegrass 2.31
Melilotus officinales Yellow sweetclover
-0.72
Medicago sativa Alfalfa 0.82
Astragalus cicer Milk vetch 1.90
Atmospheric N2 0.00
Soil N 7.00
Table 15-3 (pg. 380 of text)
-
Lifestyles of N2 fixing bacteria(diazotrophs)
Free living
Living in consortia e.g. stromatolites, soil crusts
Plant associative (living in rhizosphere)
Symbiotic
-
Middle Proterozoic formations ofthe Hakatai Shale in Grand
Canyon National Park.Lens cap is 55 mm.
Livingstromatolites
Diazotrophicbacteria
in consortia
-
Diazotrophicbacteria
in consortiasoil crusts
-
Cyanobacteria insoil crusts
-
Diazotrophicbacteria
in consortia
-
Cyanobacteria Photosynthetic and
dinitrogen fixing heterocysts separate
the two functions
Anabaena
Microcystis
Nostoc
Free-living
-
Cyanobacteria Oldest known fossils
3.5 bybp (oldest rocks are 3.8 bypb)
filamentous Palaeolyngbya
colonial chroococcalean
-
Cyanobacteria heterocysts
Heterocyst
-
Symbiotic N2-fixation: Azolla - Anabena
S. Navie
-
Symbiotic N2-fixation: Azolla - Anabena
-
Rice-Azolla-Fish, China
Azolla to feed cows, Thailand
Rice-Azolla-Ducks, KoreaTakao Furuno
Symbiotic N2-fixation: Azolla - Anabena
-
Paul Cox
Cycas micronesica
-
Cycad root nodules
-
Paul Cox
Cycas micronesica-N-methylamino-L-alanine (BMAA)
-
Guam flying fox(Pteropus mariannus)
bio-magnification
-N-methylamino-L-alanine (BMAA)
-
Flying Fox with Prunes and Cream Sauce
6 flying foxes (in case you are wondering, these are bats) 1 pound prunes 1_ cup white wine salt, pepper 1/4 cup flour 2 oz. butter 1 tbsp red currantjelly 1 cup thick cream
Remove the flesh from the flying foxes. Either plunge the animals in boilingwater for a while, then skin them and remove the flesh from the bones, orroast the animals for a little over an open fire, remove, and when cool, breakopen down the backbone and remove the flesh from the skin.
Soak the prunes overnight in 1 cup of the wine, then heat for about tenminutes in the wine before using. Season the flying fox meat with salt andpepper and roll in flour. Saute in butter over a low heat until brown. Add therest of the wine, cover and cook another 20 minutes. Add the juice from theprunes, and transfer the prunes onto a serving dish. Cook the meat in theprune juice, uncovered, for another 10 minutes, then place on the servingdish with the prunes.
The preparation of this recipe requires an ingredient which is now aprotected species.
-
Spinal cord
Amyotrophic Lateral Sclerosis-N-methylamino-L-alanine
Mimics glutamateand acts as agonistat glutamatereceptor
-
Frankia in alder root nodules
Actinorhizal symbioses
-
Frankia vesicles
Frankia root nodulesSpores &hyphae
-
ColletiaDicaria
Ceanothus
-
Ceanothus
-
Myrica faya
Native to Canary Islands
Actinorhizal root nodules
-
Invasive in Hawaii (no native N2 - fixing pioneers)
Myrica faya
-
Legumes &N-fixingbacteria
Simms
Soil-dwelling rhizobia infectlegume roots
-
Signals early in infection Complex handshaking between legume root
and rhizobium
Incorrect signal
Correctsignal
-
Legume & N-fixing bacteria Rhizobia engulfed into
nodule cells
Differentiate intobacteroids
Simms
-
Illustration: M.S. Hargrove
Photo: D. Hume
Photo and illustration: R. F. Denison
Leghemoglobin
-
RhizobiumAgrobacterium
Sawada et al. 2003
SinorhizobiumEnsifer
Mesorhizobium,
BradyrhizobiumNitrobacter, Afipia
Methylobacterium
Sinorhizobium
DevosiaAzorhizobium
Ralstonia
Burkholderia
Rickettsia
BartonellaAminobacter, Phyllobacterium
-Rhizobial symbioses have evolved ~10 times-Nested parasites & non-symbionts
Brucella
Proteobacteria
-
Symbiotic plasmid of Rhizobium etli
Vctor Gonzlez et al. Genome Biology 2003 4(6):R36
-
The nodulation genes nodABCDIJ are represented in blue
The nitrogen-fixation genes nifHDKNEXAB, fixABCX and fdxBN are represented in yellow
plasmid 42d
M. loti MAFF303099
plasmid NGR234a
M. loti MAFF303099
B. japonicum
S. meliloti pSymA
Vctor Gonzlez et al. Genome Biology 2003 4(6):R36
-
Victor Kunin et al. Genome Res. 2005; 15: 954-959
Figure 3. Three-dimensional representation of the net of life
-
Hazards of symbiotic life (or an animal dispersal agent?)
Clover Root CurculioSitona hispidula (Fabricus)