Can hsp90α-Targeted siRNA Combined With TMZ Be a Future Therapy for Glioma?
Transcript of Can hsp90α-Targeted siRNA Combined With TMZ Be a Future Therapy for Glioma?

Cancer Investigation, 28:608–614, 2010ISSN: 0735-7907 print / 1532-4192 onlineCopyright c© Informa Healthcare USA, Inc.DOI: 10.3109/07357901003630967
ORIGINAL ARTICLECellular and Molecular Biology
Can hsp90α-Targeted siRNA Combined With TMZ Bea Future Therapy for Glioma?
Nichola Cruickshanks,1 Leroy Shervington,1 Rahima Patel,1 Chinmay Munje,1 Dipti Thakkar,1
and Amal Shervington1
1Brain Tumour North West, Faculty of Science and Technology, University of Central Lancashire, Preston, United Kingdom
ABSTRACT
Hsp90α’s vital role in cell cycle progression and apoptosis together with its presence ingliomas and absence in normal tissue, make it a credible target for cancer therapy. Three setsof dsRNA oligos designed to align different regions of the hsp90α sequence were used todownregulate hsp90α. SiRNA 1, 2, and 3 resulted in significant levels of silencing of hsp90α
after 48 hr treatment (p < .0001). Concurrent treatment of the glioma cell line U87-MG withsiRNA 1 and temozolomide (TMZ) resulted in a 13-fold reduction in the dose of TMZ required toachieve a similar effect if TMZ was used alone.
INTRODUCTION
The acquisition and accumulation of mutations generally re-sult in deregulated cell proliferation and suppressed cell death(1). However, the 90-kDa heat shock protein (hsp90) can sta-bilize these mutations and counteract their effect on proteinfolding conformations that contribute to both cell proliferationand apoptosis (1, 2). Hsp90 supported by multitudes of dynamiccochaperone complexes binds to an array of client proteins andplays a vital role in the folding, stabilization, and refolding ofnewly synthesized proteins, in addition to denatured proteins(3–5). Its overexpression protects cells against apoptotic celldeath (1, 2).
Natural product derivatives such as the macrocyclic com-pound radicicol (RA) has been identified and developed to in-hibit the hsp90 protein folding machinery (6–8); however, morerecent developments have shown that hsp90 may be silenced
Keywords Glioma, Normal brain, Hsp90α, siRNACorrespondence to:Dr Amal Shervington, PhD,University of Central Lancashire, Faculty of Science,Corporation Street, Preston,Pr1 2HE United Kingdom.email: [email protected] work was supported by a grant from the Royal PrestonHospital, SeedCorn and Sydney Driscoll NeuroscienceFoundation (SDNF).
at the gene level using siRNA specifically designed to targethuman hsp90.
Humans have two major cytoplasmic isoforms, namelyhsp90α and hsp90β. Hsp90α being the major induced formwhile hsp90β the minor constitutively expressed form (9, 10).Hsp90α, owing to its inducible expression and its involvementin cell cycle progression and apoptosis, in addition to its rolein malignant phenotypes associated with invasion, angiogene-sis, and metastasis (2, 11), is also involved in tumorigenesisand the regulation of cancer (12, 13). Furthermore, its exclu-sive presence in glioma tissue and absence in brain normaltissue (14) validates its role as a possible therapeutic target forgliomas.
The aim of this study was to downregulate hsp90α in gliomaand normal human brain cell lines using hsp90α-targeted siRNAand to determine the subsequent sensitivity of the cells tochemotherapeutic agents.
MATERIALS AND METHODS
Cell lines
The human brain cell line U87-MG (glioblastoma astrocy-toma) (ECCAC, UK) was cultured in Eagle’s minimum essentialmedium (EMEM) (Sigma, UK) while the normal human astro-cytes (NHA) were cultured in astrocyte medium (ScienCell,USA).
608
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

qRT-PCR (quantitative real-time-polymerasechain reaction)
An average of 1 ρg per cell of mRNA was extractedfrom cell lines using mRNA Isolation Kit (Roche, UK)(15); 100 ng was transcribed to cDNA using a first strandcDNA Synthesis Kit (Roche, UK), which was then usedas a template for PCR. qRT-PCR was used to evaluate theexpression of hsp90α utilizing glyceraldehyde 3-phosphatedehydrogenase (GAPDH) as a control. Primer (TIB MOL-BIOL, Germany) sequence and length of the amplicons wereas follows: hsp90α (189 bp) sense—5′tctggaagatccccagacac,antisense—5′agtcatccctcagccagaga; GAPDH (238 bp)sense—5′gagtcaacggatttggtcgt, antisense—5′ttgattttggagggatc-tcg. qRT-PCR was performed in triplicate using Fast Start DNAmaster PLUS SYBR Green 1 (Roche, UK) in a LightCyclerreal-time PCR detection system (Roche Diagnostics, Germany)(15). Quantitative amplification was monitored by the levelof fluorescence reflecting the cycle number at the detectionthreshold (crossing point) using a standard curve (15).
siRNA transfections
siRNA design begins with the AUG start codon of the hsp90transcript and scan for AA dinucleotide sequences recordingeach AA and the 3′ adjacent 19 nucleotides as potential siRNAtarget sites. Generally all siRNAs with 3′ overhanging UU din-ucleotides are the most effective. Three sets of predesignedhsp90α siRNA duplexes composing of 21nt were designed us-ing a proprietary algorithm developed by Cenix BioScience andowned by Ambion (UK). Oligonucleotides used for the siRNAexperiments were as follows:
siRNA1 (NM 005348, targeted exon 5) (sense)5′GCAGGAAGAGAAAAAAACAtt3′ (antisense) 5′UGUUUUUUUCUCUUCCUGCtt3′; siRNA2 (NM 005348, targetedexon 5) (sense) 5′CGUGAUAAAGAAGUAAGCGtt3′
(antisense) 5′CGCUUACUUCUUUAUCACGtt3′; siRNA3(NM 005348, targeted exon 9) (sense) 5′GCGAUGAUGAGGCUGAAGAtt3′ (antisense) 5′UCUUCAGCCUCAUCAUCGCtt3′;GAPDH siRNA (NCBI NM 002046, targeted exons 7 and 8)(sense) 5′GGUCAUCCAUGACAACUUUdTdT3′ (antisense)5′AAAGUUGUCAUGGAUGACCdTdT3′). Negative scram-bled siRNA was used. The annealed siRNAs were resuspendedto achieve a final concentration of 20 µM and was deliveredto the cells using the siPORTTM siRNA Electroporation Kitaccording to the manufacturers protocol (15, 16). Cells were ei-ther incubated for 24 hr or 48 hr, following incubation, the cellswere harvested for mRNA isolation ready for RT-PCR, whilethe chamber slides were fixed with 4%% paraformaldyhydeand the 96-well plates were subjected to further treatment withvarious concentrations of cisplatin (0–50 µM) or TMZ (0–1000µM) (15, 16) for a further 48 hr to asses cell viability. Controlwells containing the drug and media alone were included forthe detection of possible background luminescence.
Immunofluorescence
Cells were initiated on chamber slides so as to achieve 60%confluence after 48 hr. After this time, cells were fixed with4% paraformaldehyde and washed in blocking solution beforethe addition of monoclonal antibody against primary hsp90α
(1:50) (Cambridge Bioscience, UK) (16). The primary antibod-ies were detected with goat antirat IgG FITC (1:128, Sigma,UK) and counterstained with 1.5 µg/mL VECTASHIELD R©mounting medium with Propidium Iodide (PI, USA). Cells werevisualized on an Axiovert 200M LSM 510 laser scanning con-focal microscope (Carl Zeiss Ltd, UK). For each slide, totalcells together with the positive staining cells were counted inat least 5 random microscopic fields. A total 250 cells werecounted.
Chemosensitivity assay
The sensitivity of U87-MG and NHA cells toward thechemotherapeutic drugs were tested in triplicate using CellTiter-Glo R© Luminescent Cell Viability Assay (Promega, UK) (16).Cells were plated in 96-well plates for 24 hr, after whichtime they were incubated with chemotherapeutic drugs cis-platin (Sigma-Aldrich, UK) and temozolamide (Provided byRoyal Preston Hospital, UK) at various concentrations for48 hr (16). All values are expressed as the mean ± standarddeviation, values of p < .05 were considered to be statisticallysignificant.
RESULTS
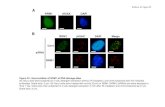
In order to assess the specificity and efficiency of silencinghsp90, predesigned siRNAs were used to effectively downreg-ulate hsp90α at the gene level.
The mRNA level of hsp90α and GAPDH were measuredusing qRT-PCR in U87-MG and NHA cell lines. siRNAs 1 and2 failed to significantly affect hsp90α after 24 hr, however, theexpression was dramatically reduced after 48 hr in U87-MGcell line (Figure 1). Although hsp90α constitutive transcriptionis downregulated in NHA, a further reduction in the transcrip-tion level was observed with the siRNA-treated cells (Figure 2).siRNA 3 demonstrated a successful downregulation of hsp90α
at 24 and 48 hr in both glioma and control cell lines (Figures 1and 2). Hsp90α protein was measured in order to correlate thetranscription to the protein level of the candidate genes aftertreatment with siRNA. While there was a reduction in the pro-tein level after 24 hr with siRNA 3 treatment, no hsp90α proteinwas detected in U87-MG after 48 hr treatment with siRNA2 and 3 (Table 1, Figure 3). In addition, no hsp90α proteinwas observed in NHA cells either before or after siRNA treat-ment. Treatment with both cisplatin and TMZ in the absence ofsiRNA demonstrated a dose-dependent decrease in cell viabil-ity. However treatment of U87-MG cells with siRNA1 2 or 3in combination with either cisplatin or TMZ was significantlymore effective in contrast to chemotherapy alone. Cisplatin IC50
(µM) for U87-MG compared to NHA cells treated with siRNA1, 2, or 3 were 15/43, 16/63, and 14/25, respectively, while TMZ
Can hsp90α-Targeted siRNA Combined With TMZ Be a Future Therapy for Glioma? 609
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

Figure 1. Expression levels of hsp90α and GAPDH in U87-MG siRNA-treated cells assessed after 24 and 48 hr, Agarose gel electrophoresisand copy numbers of gene expression (data values are mean ± standard deviation, n = 3), (A) after 24 hr (B) after 48 hr; lane 1–3 representsiRNAs 1–3 respectively, lane 4 GAPDH siRNA, lane 5 negative siRNA, and lane 6 U87-MG control cells. A statistical comparison betweenuntreated and all three experimental hsp90α siRNA-treated cells showed a significant difference in U87-MG (p = .01).
Figure 2. Expression levels of hsp90α and GAPDH in NHA siRNA-treated cells assessed after 24 and 48 hr, Agarose gel electrophoresis andcopy numbers of gene expression (data values are mean ± standard deviation, n = 3), (A) after 24 hr (B) after 48 hr; lane 1–3 represent siRNAs1–3 respectively, lane 4 GAPDH siRNA, lane 5 negative siRNA, and lane 6 NHA control cells.
610 N. Cruickshanks et al.
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

Table 1. Hsp90α Expression Quantification From siRNA-Treated Cells
Cell Line siRNA % Cell Expressing Hsp90α After Hsp90α mRNA/100 ηg Cells Extract (1 × 106 cells)24 hr 48 hr 24 hr 48 hr
Hsp90α oligo 1 71 18 11,561 17Hsp90α oligo 2 68 0 11,554 0
U87-MG Hsp90α oligo 3 0 0 0 0Negative 64 66 11,621 11,610GAPDH siRNA 73 70 11,554 11,554No siRNA 61 64 11,561 11,563
Hsp90α oligo 1 2 0 30 17Hsp90α oligo 2 0 0 20 15
NHA Hsp90α oligo 3 0 0 25 10Negative 0 0 20 30GAPDH siRNA 0 0 19 20No siRNA 0 0 20 20
IC50 (µM) were 412/454, 191/226, and 138/314, respectively.This reduction in cell viability was even more pronounced after48 hr with TMZ (200 µM and above). U87-MG cells treatedwith any of the three hsp90 siRNAs showed a significant re-duction in cell viability after 24 and 48 hr (Table 2 and Fig-ures 4 and 5), however no significant chemosensitivity was ob-served with NHA cells when treated with hsp90 siRNAs (Table2 and Figures 4 and 5). There was no significant reduction incell viability as a result of treatment with the negative controlor GAPDH siRNAs in both cell lines after a 10% reductionerror in cell viability, taking into account the application ofelectroporation (17).
DISCUSSION
Targeting hsp90α gene may be a novel therapeutic approachwith regards to glioma treatment, thus siRNA targeted towardhsp90α was designed to target the different coding regions ofthe gene. U87-MG (glioma) and NHA (normal brain control)siRNA-treated cells were tested for their chemosensitivity to-ward cisplatin and TMZ.
The results from this study showed that the expression ofhsp90α in glioma cells was reduced after 24 hr with siRNAtreatment and was completely silenced after 48-hr treatmentwith oligos 2 and 3. Since there is no constitutive hsp90α protein
Figure 3. Hsp90α protein levels assessed using immunofluorescence in siRNA-treated cells (A) U87-MG, lanes 1–6 represent siRNA 1–3,GAPDH siRNA, negative siRNA, and control cells respectively after 24-hr treatment; (B) U87-MG, lanes 1–6 represent siRNA 1–3, GAPDHsiRNA, negative siRNA, and control cells respectively after 48-hr treatment; (C) NHA, lanes 1–6 represent siRNA 1–3, GAPDH siRNA, negativesiRNA, and control cells respectively after 24-hr treatment; (D) NHA, lanes 1–6 represent siRNA 1–3, GAPDH siRNA, negative siRNA, andcontrol cells respectively after 48 hr-treatment. Cells stained with FITC conjugate secondary antibody, which bound to Hsp90α (green) antigensthe nuclei labeled with propidium iodide (red) × 63 objective magnifications.
Can hsp90α-Targeted siRNA Combined With TMZ Be a Future Therapy for Glioma? 611
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

Table 2. IC50 of two Cytotoxic Drugs in Treating Glioma Cells After 24 and 48 hr (Data Values are Mean, n = 3)
IC50 (uM)Cisplatin
Cells siRNA 1 siRNA 2 siRNA 3 Control Control/siRNA 1 Control/siRNA 2 Control/siRNA 3
24 hr 14.67 15.97 14.15 33.49 2.28 2.10 2.3748 hr 28.15 22.57 29.99 48.21 1.71 2.14 1.61
U87-MG Temozolomide24 hr 411.92 191.40 137.97 1514.63 3.68 7.91 10.9848 hr 104.93 119.11 281.94 1332.77 12.70 11.19 4.73
Cisplatin24 hr 43.09 62.54 25.11 224.45 5.21 3.59 8.9448 hr 44.71 48.03 61.96 207.08 4.63 4.31 3.34
NHA Temozolomide24 hr 453.57 226.34 313.94 686.39 1.51 3.03 2.1948 hr 606.78 785.41 726.03 967.72 1.59 1.23 1.33
in the NHA control cells, there was no siRNA silencing effectdetected.
Sequence-specific interactions between siRNA and the targetRNA are crucial and the recognition of specific sites depends
on the molecular mechanism of RNA–RNA interactions (18).Although, three sets of dsRNA oligos designed to align differentregions of hsp90α sequence were used, hsp90α-targeted siRNAs1, 2, and 3 showed significant reduction in hsp90α mRNA and
Figure 4. Cell viability assessments of NHA (A and B) and U87-MG (C and D) cells treated with increasing concentrations of cisplatin (A and C)or temozolomide (B and D) for 24 hr alone and in combination (with siRNA 1–3, GAPDH siRNA, or negative control siRNA followed by cisplatinor temozolomide). (Data values are mean ± standard deviation, n = 3).
612 N. Cruickshanks et al.
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

Figure 5. Cell viability assessments of NHA (A and B) and U87-MG (C and D) cells treated with increasing concentrations of cisplatin (A and C)or temozolomide (B and D) for 48 hr alone and in combination (with siRNA 1–3, GAPDH siRNA, or negative control siRNA followed by cisplatinor temozolomide). (Data values are mean ± standard deviation, n = 3).
protein levels after 48 hr; however, only hsp90α siRNA3 showedan effect after 24 hr.
In all the glioma cell lines examined, the expression levelof hsp90α was high in comparison to the normal brain NHAcells (14). The general hypoxic microenvironment of a tumorcell and its ability to tolerate usually lethal mutations accountsfor an increase in hsp90 and subsequently contributes to bothcell proliferation and apoptosis (19), thus the downregulated orsilenced hsp90α expression should enhance chemosensitivityand cell death.
Although cell death can be achieved by apoptosis (type I-programmed cell death) it can also be achieved by necrosis, mi-totic catastrophe, and autophagy. Autophagy has been reportedto initiate cell death in response to intracellular damage causedby hypoxia, chemotherapeutic agents, and others (20–22).
Hsp90 is an antiapoptotic that causes chemoresistance in tu-mor cells (23) and inhibits transcription using siRNA whichenhances induced cell death in glioma cell lines. Several po-tential common targets in apoptosis and autophagy resistance
pathways such as mTOR and Akt, have been identified andinhibitors of such targets could possibly increase the level ofchemosensitivity (20–22).
Exposure of U87-MG and NHA cells to increasing concen-trations of the chemotherapeutic drugs, cisplatin and TMZ, re-sults in a dose-dependent decrease in cell viability. When eithercisplatin or TMZ was combined with siRNA a slightly enhancedchemosensitivity of NHA-treated cells was detected, in contrast,after 48 hr, TMZ treatment was more effective with U87-MGsiRNA-treated cells. Tumor hsp90 is complexed with cofactorssuch as p23 and hsp70 that provides a more suitable conforma-tion that binds with inhibitors (20). As a result, unlike tumorcells, normal cells are not sensitive to hsp90 inhibitors (24).
Although siRNA 1 and 2 showed a belated hsp90α downreg-ulation, increased chemosensitivity was observed with gliomacells treated with a combination of siRNA1 or siRNA2 and TMZshowing a 13- and 11-fold reduction, respectively, in the con-centration of TMZ required in order to achieve the same toxiceffect.
Can hsp90α-Targeted siRNA Combined With TMZ Be a Future Therapy for Glioma? 613
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.

Interestingly, oligo 1 and 2 which targets exon 5 showed abelated downregulation compared with oligo 3 which targetshsp90α exon 9, suggesting that the proximity of the oligo to theinitiation site for translation enhances the efficiency of silenc-ing and consequently cell chemosensitivity. Analysis of siRNA-duplex sequences shows that the thermodynamic stability oftheir two RNA ends determines which strand is incorporatedinto RNA-induced silencing complex (RISC) (25). Althoughthese siRNA have been designed by Ambion using algorithmsand if the thermodynamics of siRNA duplexes and their incor-porations into RISCs were the only factors for effective RNAiin mammalian cells, then all the three siRNA should cleavetheir target mRNAs with high efficiencies. However, not allsiRISCs that have been efficiently loaded with antisense strandsare equally efficient in silencing target genes and therefore, it isimportant to further our understanding of the mechanisms thatare involved in the control of silencing gene expression fromany target mRNA in order to decrease off-target effects and todefine physiological roles for the RISC-enzyme complexes.
The data suggest that combining both siRNA and TMZ en-hances the sensitivity of the alkylating agent, resulting in a sig-nificant reduction in concentration of TMZ required to achievethe same effect while reducing side effects and improving thelife expectancy for patients.
CONCLUSION
The specific inhibition of hsp90α expression by siRNA maybe a favorable therapeutic approach compared with conventionalchemotherapies, due to its target specificity and reduced toxicity.This study supports the hypothesis that antisense oligodeoxynu-cleotides directed against hsp90α can inhibit hsp90α directly,leading to enhance chemosensitivity. Combination treatmentwas significantly more effective, suggesting that either com-bination of siRNA 1 or 2 followed by TMZ (200–400 µM) after48 hr could possibily be an effective form of therapy.
Declaration of Interest: The authors report no conflict ofinterest. The authors alone are responsible for the content andwriting of this paper.
REFERENCES1. Mosser, D.D.; Morimoto, R.I. Molecular chaperones and the stress
of oncogenesis. Oncogene 2004, 23, 2907–2929.2. Graner, M.; Bigner, D. Chaperone proteins and brain tumours:
potential targets and possible therapeutics. Neuro-Oncol 2005, 7,260–277.
3. Pearl, L.H.; Prodromou, G. Structure, function, and mechanismof the Hsp90 molecular chaperone. Adv Protein Chem 2001, 59,157–186.
4. Picard, D. Heat-shock protein 90, a chaperone for folding and reg-ulation. Cell Mol Life Sci 2002, 59, 1640–1648.
5. Pratt, W.B.; Toft, D.O. Regulation of signalling protein function andtrafficking by the hsp90/hsp70-based chaperone machinery. ExpBiol Med 2003, 228, 111–133.
6. Goetz, M.P.; Toft, D.O.; Ames, M.M.; Erlichman, C. The Hsp90chaperone complex as a novel target for cancer therapy. Ann Oncol2003, 14, 1169–1176.
7. Workman, P.; Powers, M. Chaperoning cell death: a critical dualrole for Hsp90 in small-cell lung cancer. Nature Chemical Biol2007, 3, 455–457.
8. Hadden, M.K.; Lubbers, D.J.; Blagg, B.S. Geldanamycin, radicicol,and chimeric inhibitors of the Hsp90 N-terminal ATP binding site.Curr Top Med Chem 2006, 6, 1173–1182.
9. Sreedhar, A.; Kalmar, E.; Csermely, P.; Shen, Y. Hsp90 isoforms:functions, expression and clinical importance. FEBS 2004, 562,11–15.
10. Pearl, L.H.; Prodromou, G.; Workman, P. The Hsp90 molecularchaperone: an open and shut case for treatment. Biochem J 2008,410, 439–453.
11. Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer:diagnosis, prognostic predictive and treatment implications. CellStress Chaperones 2005, 10, 86–103.
12. Eustace, B.K.; Sakurai, T.; Stewart, T.; Yimlamai, D.; Unger, C.;Zehetmeier, C.; Lain, B.; Torella, C.; Henning, S.W.; Beste, G.;Scroggins, B.T.; Neckers, L.; Ilag, L.; Jay, D.G. Functional pro-teomic screens reveal an essential extracellular role for hsp90 al-pha in cancer cell invasiveness. Nat Cell Biol 2004, 6, 507–514.
13. Chen, X.; Zhang, Y.; Wang, J.; Li, X.; Cheng, X.; Zhang, Y.; Wu, N.;Shen, Y. Diverse effects of Stat1 on the regulation of hsp90 geneunder heat shock. J Cell Biochem 2007, 102, 1059–1066.
14. Shervington, A.; Cruickshanks, N.; Lea, R.; Roberts, G.; Dawson,T.; Shervington, L. Can the lack of HSP90α protein in brain normaltissue and cell lines, rationalise it as a possible therapeutic targetfor gliomas? Cancer Invest 2008, 26, 900–904.
15. Patel, R.; Shervington, L.; Lea, R.; Shervington, A. Epigenetic si-lencing of telomerase and a non-alkylating agent as a novel ther-apeutic approach for glioma. Brain Res 2007, 1188, 173–181.
16. Shervington, A.; Lu, C. Expression of multidrug resistance genesin normal and cancer stem cells. Cancer Invest 2008, 26, 535–542.
17. Jarvis, R.; King, A. Optimizing chemical transfection and electro-poration of siRNAs. Ambion TechNotes 2003, 10, 12–15.
18. Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.;Ttuschl, T. Functional anatomy of siRNAs for mediating efficientRNAi in Drosophila melanogaster embryo lysate. EMBO J 2001,20, 6877–6888.
19. Whitesell, L.; Lindquist, S. Hsp90 and the chaperoning of cancer.Nature Reviews: Cancer 2005, 5, 761–772.
20. Lefranc, F.; Facchini, V.; Kiss, R. Proautophagic drugs: a novelmeans to combat apoptosis-resistant cancers, with a Special Em-phasis on Glioblastomas. The Oncologist 2007, 12, 1395–1403.
21. Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.;Thompson, C.B. Growth factor regulation of autophagy and cellsurvival in the absence of apoptosis. Cell 2005, 120, 237–248.
22. Kanzawa, T.; Germano, I.M.; Komata, T.; Ito, H.; Kondo, Y.; Kondo,S. Role of autophagy in temozolomide-induced cytotoxicity for ma-lignant glioma cells. Cell Death Differ 2004, 11, 448–457.
23. Cervantes-Gomez, F.; Nimmanapalli, R.; Gandhi, V. Transcriptioninhibition of heat shock proteins: a strategy for combination of 17-allylamino-17-demethoxygeldanamycin and Actinomycin D. Can-cer Res 2009, 1, 3947–3954.
24. Workman, P. Altered states: selectively drugging the Hsp90 cancerchaperone. Trends Mol Med 2003, 10, 47–51.
25. Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs andmiRNAs exhibit strand bias. Cell 2003, 115, 209–216.
614 N. Cruickshanks et al.
Can
cer
Inve
st D
ownl
oade
d fr
om in
form
ahea
lthca
re.c
om b
y U
nive
rsity
of
Ade
laid
e on
11/
16/1
4Fo
r pe
rson
al u
se o
nly.