Nucleotide sequence of the 5′-flanking region of the mouse κ-FGF oncogene exhibits an alternating...
Transcript of Nucleotide sequence of the 5′-flanking region of the mouse κ-FGF oncogene exhibits an alternating...
Gene, 96 (1990) 311-312 Elsevier 311
GENE 03797
Brief Note
Nucleotide sequence of the 5'-flanking region of the mouse k-FGF oncogene exhibits an alternating purine" pyrimidine motif with the potential to form Z - D N A
(Recombinant DNA; transcription factors AP-2 and Spl; consensus binding sequences)
Jay Tiesman and Angie Rizzino
Eppley Institute for Resea,~h in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE 68198-6805 (U.S.A.)
Received by J. Marmur: 6 August 1990 Accepted: 20 August 1990
SUMMARY
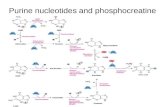
The nucleotide sequence of the 5'-flanking region of the mouse k-FGF oncogene has been determined. This sequence extends 2. lkb upstream from the transcriptional start point (tsp) and includes two Spl and two AP-2 cousen3us binding sequences inm~ediately 5' of the TATA box. In addition, the sequence contains an alternating purine: pyrimidine motif that lies approx. 1 kb upstream from the tsp.
The k-FGF oncogene (also known as hst) encodes a protein with properties similar to members of the FGF family of grov,¢h factors, k-FGF is expressed in human and mouse embryonal carcinoma cell lines, but their differen- tiation drastically reduces k-FGF expression (Rizzino et al., 1988; Ticsman and Rizzino, 1989). To gain an under- standing of the re~lation of this oncogene, we have se- quenced DNA 5' to the mouse k-FGF gene.
The genomic cosmid clone, COS I6 (Brookes et al., 1989), contains the mouse k-FGF oncogene as well over 20 kb of upstream sequence. Using subclones of this cos- mid, we have sequenced 2.1 kb of this 5' DNA and have identified two Spl (Briggs et al., 1986) and two AP-2 (Imagawa et al., 1987) consensus binding motifs that lie within 200 bp of the TATA box. Interestingly, these se- quence motifs are conserved in the human k-FGF gene sequence (Yoshida et al., 1987), despite that the 5'-flanking sequences of mouse and human k-FGF ~xhibit less than 20% sequence similarity in this 2.1-kb region.
Corre.~'pondence to: Dr. A. Rizzino, Eppley Institute, University of Nebraska Medical Center, 600 S. 42nd Street, Omaha, NE 68198-6805 (U.S.A.) Tel. (402)559-6338; Fax (402)559-4651.
Abbreviations: ARY, alternating pufine:pyrimidine; bp, base pair(s); FGF, fibroblast growth factor; kb, kilobase(s) or 1000 bp; R, A or G; tsp, transcription start point; Y, C o.r T.
In addition to the Spl and AP-2 sequence motifs, we have identified a nearly-perfect 90-bp ARY motif that lies 1 kb upstream from the tsp. This motif is not present in the human k-FGF flanking sequence. There is evidence that ARY sequences have the potential to form Z-DNA under physiological conditions (Haniford and Pulleybank, 1983; Jaworski ~t aJ., 1987) and that Z-DNA may play a role in the regulation of gene transcription (Naylor and Clark, 1990; Hamada et al., D84). The significance of this poten- tial Z-DNA forming motif is currently unclear. However, it has been hypothesized that non-B-DNA structures may play a role in allowing regulatory proteins to interact with DNA and thereby regulate gene transcription.
ACKNOWLEDGMENTS
Dr. Gordon Peters is thanked for his gift ~f cosmid COS16 and Dr. James Shull is thanked for reading this manuscript. This work was supported by grants from the National Institute of Child Health and Human Develop- ment (HD21568), the Council of Tobacco Research (2520), the National Cancer Institute (Laboratory Cancer Research Center Support Grant, CA36727), and the American Can- cer Society (ACS SIG-16). Jay Tiesman was supported by a fellowship from the Nebraska Governor's Research hfi- tiative in Biotechnology.
0378-1119/90/$03.50 © 1990 Elsevier Science Publishers B.V. (Biomedical Division)
312
TCTAGACCAGGC -2101
TGG~CTTGAA~T~AGAAATCCGC~TCCCTCTGCCTCCCAAGTG~TGGGATTAAAGGTGTGCGCCACCACTGCCCGGCGTTGTTGTTTTTCATTTTG~GAC -2001
AGGATCTTATTTTGTAGCCCAGGCTGACTTGGAACTCAGGCCAGCCTCAAATTCAGAGATACACCTGCCTCTGCCTCCTTAGTGATGGGATTAAAGGCGT -1001
GTGCCACCACTGCCCAGCCACAATAAGATCCTTTAAAGGTGTGAATTGGCTGCAAGTGGAGTGAGTGAAGGCTCACAGGCTTGGGGAGACTGAGGTCCTA -1801
GGGCAGCCTAAGGTGGAGGAGTCCTGAG~CAAAAGATCTCTGGATCTT~CCTGCAGGGAAAGGG~TGGAAACTGGACAGGCATGAGGTCCGGGGTCTGGT -1701
GGCAGACTGGGGTTAGTCCCGGAGAGC~CTA~CCT~ACCCGTGCTCTCATMYCT~CCCAACACTTCTGAACTTGGAGGAGCCGGAAGCTGGAGGTTCTG -1601
GAGC~GCAG~TGGAG~AAAAAGCAGGCCCCAAAGCAGTTTGATGATAGTGCCCAA~CTTGGTC~TCCTGACTACTAACCACTGT~AAGAG~ACC~AC~G -1501
GGTGTGGTGGCCAGCAG•TCCTTCTGCAGTCACAGAGTCAGGGATGCACT•CCAAATTGAGTTGGGGTGGGGGGACCTGCCTACCACCAGGGCAATGATC -14U1
TA~ATCTGATC~AG~CT~GC~AGCC~CA~AAAG~AA~AC~AG~GTGC~A~A~TAA~CCT~CA~CC~TC~TCCA~TGACA~A~CA~MCTAG~ -13U1
AAC~TGTTA~AGGAAGGGATGTTTCTCAGCAGG~TGTGTGTATGCATGCTTATGTGTGTACAT~ATGAGTGTGTGTGTGTGTGTG~TGTGTG~GCG~ -1201
G~GTGTGT~G~GTG~CA~ACGTG~GTGGGAG~TGTGCACAG~TGA~TTTG~T~TAGcAAG~AA~TCA~C~GCTCTACCTGCC~TGGAGGA -110~
TGCGGTTATCC•AGGCTGCCAGGTGCATCAGAATGCCTCATTAAGGGATTTGGAGACATGGTGTAATCCCCACTAAGGGATAGATAGATGCTCAAGTGTC -1001
TC~TCTAGGTCTCAGGA~CTGGCTGCATTTCCGTTGGTGTTCTGAGTA~GACCAGGAATGATTGGTGCAAGCCC~ATGACAGAGGTTTGGCAGAAGGAAG -901
c~A~AG~T~AGAAG~T~TGGT~AAATTACTGATT~T~GGTGAAGG~GTG~AG~AGG~TCYT~GG~T~T~ATcAGTGG~AGGT~A~TG~TGG~ -801
TC•AGGTC•TCAGATACCACAGAACAGGGACATTCACACCTAGGAGAACAGTA•CAGGGTT•TGGCCCAGTT•ACCC•CATCAGGCCAGAATGGCACAGT -701
TGGGTGTGGGGGTG•TGGTGGGGGAAGGGACCCTT•CAGATCCTTAGAGTTTGA•ATGCT••TGGGTTGGGAAGTTCTGG•CTCCACACTTGAAAATCT• -601
TGGGGAACTGATGC•CAATGT•TTGCCAG•TTT•TGTCCTTG•CTTGGGTTAC•TAGGAGGAAAGTG•GGGT•TCTGC1GTCCCTGAATGTCCTCAACCT -501
GAGTT•T•TTAGAAG•ACCCACTCCGTAGTTTAGACTTCCCATCACCTGGAAGACCCTTAAATTTCCAAGCCTCTGGGGCCAGACCAAGCCTTGCCTCCC -401
TCC~TC~ATC~TG~CCTACTCC~GGCTCC~TTAGAGTTACAC~CTGATAGC~AAGCCACAGTGTGACTTGCCTCCCACCGTCAG~TCAAGCCAGTC~TT -301 Spl
GGCGCAGCAGAAAGGTT•TGGCGGTTCACCAAGTGTCCCGTAAGGAAGGA•CA•AGGAGATGCCCTGGGGAGCAGAGAGCCAAGG•GGC•GGATCAACAGG -201 Spl AP-2 AP-2
TT•GAGTGCGGGGCGGAGCCAAGAGTAAGGGGTTGGGGTCTCTCCAGG•GACAGTAGCCACCGCCAGGCCCGCGCCT•CTCCCCCGGCGG•GATTGGCAG -101
GCGGCCTGCGCCCCGG•TCCCAGG•GACCGACGCCCCGCGGGGCAGGCGAGTAGGAGGGGGCGCCGGCTATATA.[ACCACTGCTCCGGAGGGCT••GCGC -1
Fig. 1. Sequence of the 5' -flanking region of the mouse k.FGF encogene. Nucleotides are numbered from the tsp ( + 1). The 9O-bp ARY sequence is boxed. The TATA box is in bold print. Spl and AP-2 binding consensus sequence motifs are underlined. Overlapping subclones of the COS16 cosmid were sequenced on both strands using the dideoxy sequencing method of Sanger et al. (1977). The EMBL accession number of this sequence is X54053.
REFERENCES
Briggs, M.R., Kadonaga, 3.T,, Bell, S.P. and Tijan, R.: Purification and biological characterization of the promoter-specific transcription fac- tor Spl. Science 234 (1986) 47-52.
Brookes, S., Smith, R., Thurlow, J., Dickson, C. and Peters, G.: The mouse homologue ofhst/k-FGF: sequence, genome organization and location relative to int-2. Nucleic Acids Res. 17 (1989) 4037-4045.
Hamada, H., Seidman, M., Howard, B.H. and German, C.M.: Enhanced gone cxpw~:ion by the poly(dT-dG)-poly(dC.dA) sequence. Mo]. Cell. Biol. 4 (1984) 2622-2630.
Haniford, D.B. and PuUeybank, D.E.: Facile transition of poly[d(TG)- d(CA)] into a leit-handed helix in physiological conditions. Nature 302 (1983) 632-634.
Jaworski, A., Hsieh, W.-T., Blaho, J.A., Larson, J.E. and Wells, R.D.: LeR-handed DNA in vivo. Science 238 (1987) 773-777.
Imagawa, M., Chiu, R. anti Karin, M.: Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell 51 (1987) 251-260.
Naylor, L.H. and Clark, E.M.: d(TG),-d(CA), sequences upstream ofthe rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids .~.e~.. 18 (1990) 1595-1601.
Rizzino, A., Kuszynski, C., Ruff, E. and Tiesman, .I.: Production and utilization of growth factors related to fibroblast growth factor by embryonal carcinoma cells and their differentiated cells. Develop. Biol. 129 (1988) 61-71.
Sanger, F., Nicklen, S. and Coulson, A.R.: DNA sequencing with chain- terminating inhibitors. Prec. Natl. Acad. Sci. USA 74 (1977) 5463 -5467.
Tiesman, J. and Rizzino, A.: Expression and developmental regulation of the k-FGF oncogene in human and murine embryonal carcinoma cells. In Vitro Cell. Develop. Biol. 25 (1989) 1193-1198.
Yoshida, T., Miyagawa, K., Odagiri, H., Sakamoto, H., Little, P.F.R., Terada, M. and Sugimura~ T.: Genomic sequence of hst, a trans- forming gene encoding a protein homologous to fibroblast growth factors ar, d the int-2-encoded protein. Prec. Natl. Acad. Sci. USA 84 (1987) 7305-7309.