A B Cornelissen FigS2 - Genes & Developmentgenesdev.cshlp.org/content/suppl/2019/12/23/gad...DIP...
Transcript of A B Cornelissen FigS2 - Genes & Developmentgenesdev.cshlp.org/content/suppl/2019/12/23/gad...DIP...

phospho-STAT5
total-STAT5
β-actin
β-actin
DIP mixFull medium
DMSO300nM TSA - + - +
+ - + -+ + - -- - + +
Histone H3
TRPS1
Cytoplasmic
HDAC1
CHD4
GAPDH
HDAC2
RBBP7
TOP1
Membranous Soluble-nuclear Chromatin-bound
sgTr
ps1
#1 +
EV
sgTr
ps1
#1 +
TRP
S1tru
nc
sgSc
r #1
+ EV
sgTr
ps1
#1 +
EV
sgTr
ps1
#1 +
TRP
S1tru
nc
sgSc
r #1
+ EV
sgTr
ps1
#1 +
EV
sgTr
ps1
#1 +
TRP
S1tru
nc
sgSc
r #1
+ EV
sgTr
ps1
#1 +
EV
sgTr
ps1
#1 +
TRP
S1tru
nc
sgSc
r #1
+ EV
FL
TR
B
C
F G
2 3 4
887aa686aa309aaZinc fingerNLS
0.0
0.5
1.0
Full mediumDIP mixTSA (nM)
---+++
300800
Rel
ativ
e β-
case
in e
xpre
ssio
n(n
orm
aliz
ed to
β-a
ctin
)
-+0
***
1 2 3 4 5 6
GATATGGCCTGCACCCCATCAGG CCTCCGAGGGTGTGAAACCTTCAClones #1 + #3Clone #2
A Cornelissen_FigS2
HDAC1
β-actin
TRPS1trunc - + - +EV + - + -
sgScr #1 sgTrps1 #1
β-actin
HDAC2
TRPS1FL
TR
TRPS1trunc - + - +EV + - + -
sgScr #1 sgTrps1 #1D
E
0 50 100 150 2000
50
100
sgScr #1 + EVsgScr #1 + TRPS1trunc
sgTrps1 #1 + EVsgTrps1 #1 + TRPS1trunc
Time (hours)
Con
fluen
cy (%
)
TRPS1
β-actin
shNT shTrps1 #2
0 20 40 60 80 1000
20
40
60
80
Time (hours)
shNT
shTrps1 #2
Con
fluen
cy (%
)
0.0
0.5
1.0
1.5
shNT
shTrps1 #2
DIP mix
-+-++-+-
++--
Rel
ativ
e β-
case
in e
xpre
ssio
n(n
orm
aliz
ed to
β-a
ctin
)
**
0.0
0.5
1.0
1.5
2.0
Rel
ativ
e β-
case
in e
xpre
ssio
n(n
orm
aliz
ed to
β-a
ctin
)
EVTRPS1trunc
DIP mix
-+
+sgTrps1 #1
+-
+
-+
+sgScr #1
+-
+
+-
-sgScr
#1
+-
-sgTrps1
#1
ns
ns
H
I
J K
LsgScr #1 sgTrps1 #1
EVTRPS1trunc - +
+ -- ++ -
TRPS1
β-actin
FL
TR
phospho-STAT5
β-actin
#1 #2 #1 #2 #3
sgScr sgTrps1
total-STAT5
β-actin
TRPS1
#1 #2 #1 #3
sgScr sgTrps1
DIP mix + + + + + - - - -
M N
0
2
4
6
8
10
Maml Mkl1 Tcf3 Hoxd9 Klf3 Gata3(neg region)
HDAC2 ChIP
sgScr #1 + mTRPS1truncsgScr #1 + EV
sgTrps1 #1 + mTRPS1truncsgTrps1 #1 + EV
Fold
enr
ichm
ent
ns***
ns
ns**
ns
ns***
ns
ns***
ns
ns***
nsns
ns
ns
0
5
10
15
20
Fold
enr
ichm
ent
Maml Mkl1 Tcf3 Hoxd9 Klf3 Gata3(neg region)
HDAC1 ChIP
sgScr #1 + mTRPS1truncsgScr #1 + EV
sgTrps1 #1 + mTRPS1truncsgTrps1 #1 + EV
ns***
ns
ns*
ns
ns***
nsns
ns
ns
ns***
ns
ns
ns
ns

3
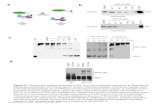
Supplementary Figure S2. Loss of TRPS1 affects HDAC1 and HDAC2 DNA binding and lactogenic
differentiation, while TRPS1trunc expression is dispensable. (A) Schematic representation of the Trps1
gene locus with the locations of the guide RNAs used to target Trps1 indicated. PAM sequences are
indicated in red. Numbered boxes represent exons of the canonical gene transcript. (B) Schematic
representation of TRPS1trunc cDNA reflecting loss of exon 5 and 6. Numbered boxes represent exons of
the canonical gene transcript. (C) Subcellular fractionation of HC11 cells with expression full-length
TRPS1, TRPS1 deficient or expression of TRPS1trunc. NuRD complex members are indicated with a green
lining. GAPDH was used as a marker for the cytoplasmic fraction, Topoisomerase 1 (TOP1) for the
soluble-nuclear fraction and histone H3 for the chromatin-bound fraction. Black arrowhead, full-length
TRPS1 (FL); grey arrowhead, truncated TRPS1 (TR). (D) Western blot analysis of HDAC1 and HDAC2
expression in HC11 control and TRPS1 knockout cells transduced with a control or TRPS1trunc construct.
β-actin was used as a loading control. Black arrowhead, full-length TRPS1 (FL); grey arrowhead,
truncated TRPS1 (TR). (E) HDAC1 and HDAC2 ChIP-qPCR analysis in HC11 control and TRPS1
knockout cells transduced with a control or TRPS1trunc construct. Five target regions and one negative
control region are shown. Data represent mean+standard deviation (SD). Two-way ANOVA: *** p<0.001,
** p<0.01, * p<0.05, ns p>0.05. (F) Western blot analysis of phospho-STAT5 and total-STAT5 expression
in HC11 cells treated with DIP mix compared to full medium, in combination with DMSO or 300nM
Trichostatin A (TSA). (G) RT-qPCR analysis for β-casein (Csn2) expression in HC11 cells treated with
DIP mix, in combination with DMSO, 80nM or 300nM TSA. Data represent mean+SD, n=2. One-way
ANOVA: ** p<0.01, * p<0.05. (H) Western blot analysis of phospho-STAT5 and total-STAT5 expression in
HC11 control and TRPS1 knockout cells treated with or without DIP mix. β-actin was used as a loading
control. (I) Western blot analysis of TRPS1 expression in HC11 cells transduced with a non-targeting
shRNA or shRNA #2 targeting Trps1 (as indicated in Figure S1B). β-actin was used as a loading control.
(J) Cell proliferation of HC11 cells transduced with a non-targeting shRNA or an shRNA targeting Trps1,
as quantified using IncuCyte imaging for 100 hours. Data represent mean±SEM. (K) RT-qPCR analysis of
β-casein (Csn2) expression levels in HC11 cells transduced with a non-targeting shRNA or targeting
Trps1 upon treatment with DIP mix. Data represent mean+SD, n=3. T-test: ** p<0.01. (L) Western blot
analysis of TRPS1 expression in HC11 control and TRPS1 knockout cells transduced with a control or

4
TRPS1trunc construct. β-actin was used as a loading control. Black arrowhead, full-length TRPS1 (FL);
grey arrowhead, truncated TRPS1 (TR). (M) Cell proliferation of HC11 control and TRPS1 knockout cells
transduced with a control or TRPS1trunc construct, as quantified using IncuCyte imaging for 200 hours.
Data represent mean±SEM. (N) RT-qPCR analysis of β-casein (Csn2) expression in HC11 control and
TRPS1 knockout cells transduced with a control or TRPS1trunc construct upon treatment with DIP mix.
Data represent mean+SD, n=2. One-way ANOVA: ns p>0.05.