Supplemental Figure 1 - jbc.org€¦ · Supplemental Figure 5. Expression of epiprofin/Sp6...
Transcript of Supplemental Figure 1 - jbc.org€¦ · Supplemental Figure 5. Expression of epiprofin/Sp6...

dental pulp SP MP
Oil-
Red-
O
cont
rol
adip
ogen
ic co
nd.
A B
dental pulp SP MP
cont
rol
neur
ogen
ic co
nd.
C
- + - + - + dental pulp SP MP
PPARγ Hprt
D
Rela
tive
expr
essio
n
40
0
30
10
20
50 PPARγ/GAPDH
- + - + - + dental pulp SP MP
Rela
tive
expr
essio
n
100
0
75
25
50
125 Neurofilament-M/β-actin
- + - + - + dental pulp SP MP
*
*
Supplemental Figure 1

rat dental epithelium (SF2 cells)
pEF6/GFP-PDGFtm-myc-HA
SF2/GFP cells (expressing GFP-myc-HA tag)
selection
co-cultured with SP cells
GFP
phase (SF2)
SF2/GFP
SP
Supplemental Figure 2

A B Ambn
CK14
Klf4
Sox2
Oct3/4
Nanog
GAPDH
SF2-24 iPS
GAPDH
Ambn 7 12 13 17 19 24 Clone No.
Rat dental epithelium (SF2 cell)
Supplemental Figure 3

A B
MEF iPS tooth germ
mAmbn GAPDH
m total-RNA
GAPDH
rAmbn
0 1 10 100 120 150 200 (ng)
200 150 120 100 10 0 1 (ng) r total-RNA
mAmbn
Supplemental Figure 4

Supplemental Figure 5
Ambn
GAPDH
Epiprofin/Sp6
1 7 10 (days)
SF2-24+iPS

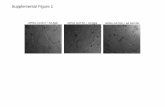
SUPPLEMENTAL INFORMATION Supplemental Figure 1. Adipogenic and neurogenic induction of SP cells. (A) Adipocyte-specific Oil-Red-O staining of dental pulp, SP, and MP cells in regular or adipogenic induction culture (adipogenic cond.) medium. (B) RT-PCR and real-time PCR analyses of the adipocyte marker gene PPARγ in dental pulp, SP, and MP cells in adipogenic induction culture medium. (C) Phase images of dental pulp, SP and MP cells cultured in regular or neurogenic induction culture (neurogenic cond.) medium. (D) Western blotting of dental pulp, SP, and MP cells cultured in regular (-) or neurogenic induction culture (+) medium was performed using the anti-Neurofilament-M antibody. The relative intensities of bands obtained with western blots were scanned and quantified using an LAS UV4000 system (Fujifilm-GE Healthcare, Tokyo, Japan). Supplemental Figure 2. Schematic diagram of establishment of SF2 cells expressing PDGFtm-GFP-myc-HA. Rat-derived dental epithelial SF2 cells were transfected with a GFP-PDGFtm-myc-HA expression vector and selected with antibiotics. GFP in the SF2 cells was visualized using fluorescence microscopy. SF2/GFP cells were co-cultured with SP cells (bottom) and separated with the anti-HA antibody. Supplemental Figure 3. Gene expression of subcloned SF2 cell lines and iPS cells. (A) Ambn expression in dental epithelial cells subcloned from SF2 cells. Clone SF2-24 expressed Ambn at the highest level and was used for co-culturing with iPS cells. (B) SF2-24 cells expressed the ameloblast marker genes CK14 and Ambn, but not stem cell markers. In contrast, stem cell markers were observed in iPS cells, but not ameloblast markers. Supplemental Figure 4. Design of mouse-specific Ambn primer. Mouse and rat Ambn mRNA showed a high degree of homology (94.2%), thus regular Ambn primers for RT-PCR are able to precisely distinguish between them. To resolve this problem, we employed a mouse Ambn LNA primer. The accuracy of the mouse-specific Ambn LNA primer was confirmed by PCR with gradient cDNA mixtures containing serial dilution ratios of mouse and/or rat cDNAs. (A) The expected size of the amplicon using the mouse LNA primer was observed only with mouse cDNA, while the intensity of the band increased in a cDNA dose-dependent manner. (B) Undifferentiated iPS cells and MEF feeder cells did not express Ambn mRNA. A mouse tooth germ sample served as a positive control for the mouse LNA primer. Supplemental Figure 5. Expression of epiprofin/Sp6 transcription factor in iPS cells co-cultured with SF2-24 cells. Time course analyses of Ambn and epiprofin/Sp6 gene expressions in iPS cells co-cultured with SF2-24 cells for 1 (1D), 7 (7D) and 10 days (10D) were analyzed by RT-PCR.

Supplemental Table1
Gene name forward primer reverse primer Size(bp)
mouse/rat Ambn 5‘-GCGTTTCCAAGAGCCCTGATAAC-3’ 5‘-AAGAAGCAGTGTCACATTTCCTGG-3’ 366/368
mouse/rat CK14 5‘-GAGAAGAACCGCAAGGATGC-3’ 5‘-AGGTTATTCTCCAGGGATGCTTTC-3’ 191
mouse/rat GAPDH 5‘-GGAGCGAGACCCCACTAACATC-3’ 5‘-CTCGTGGTTCACACCCATCAC-3’ 181
mouse Ambn 5‘-GCAGGTGGCACCATCCGALA-3’ 5‘-AAGAAGCAGTGTCACATTTCCTGG-3’ 1104
rat Ambn 5‘-GCTACCCTTCCACAGGGAAGGTLA-3’ 5‘-AAGAAGCGGTGTCACATTTCCTGG-3’ 267
mouse Enam 5‘-GGGGACCACCAACAGCGTTTGGACGGCCA-3’ 5‘-CCCGGAGGTAGGAGAGGGAGGTTTACC-3’ 614
rat Enam 5‘-CAGGACCACCAACAGGGTTCGGACGA-3’ 5‘-TCTTGAGGTAGGAGGGGAAGGTTTGT-3’ 626
mouse CK14 5‘-GGCTGCCGATGACTTCCGGAC-3’ 5‘-CAGCAGTATCTGCGTCCACGCA-3’ 909
rat CK14 5‘-AGACTACAGCCCCTACTTCAAGACC-3’ 5‘-AGGTTATTCTCCAGGGATGCTTTC-3’ 597
mouse p63 5‘-GGCATTTCAGCACTATTTAGGTGG-3’ 5‘-CACTGGTGTGAGGAGACAAACTGTG-3’ 110
mouse Epiprofin 5‘-TCAGCCTGCTTTGGGAGGATAC-3’ 5‘-ATGCTTCTTCTTGCCCCCATCG-3’ 313
Klf4 5‘-CACCATGGACCCGGGCGTGGCTGCCAGAAA-3’ 5‘-TTAGGCTGTTCTTTTCCGGGGCCACGA-3’ 739
Sox2 5‘-GGTTACCTCTTCCTCCCACTCCAG-3’ 5‘-TCACATGTGCGACAGGGGCAG-3’ 193
Oct3/4 5‘-TCTTTCCACCAGGCCCCCGGCTC-3’ 5‘-TGCGGGCGGACATGGGGAGATCC-3’ 224
Nanog 5‘-CAGGTGTTTGAGGGTAGCTC-3’ 5‘-CGGTTCATCATGGTACAGTC-3’ 223
Fgf4 5‘-CGTGGTGAGCATCTTCGGAGTGG-3’ 5‘-CCTTCTTGGTCCGCCCGTTCTTA-3’ 197
Gdf3 5‘-GTTCCAACCTGTGCCTCGCGTCTT-3’ 5‘-AGCGAGGCATGGAGAGAGCGGAGCAG-3’ 570
Cdx2 5‘-GGCGAAACCTGTGCGAGTGGATGCGGAA-3’ 5‘-GATTGCTGTGCCGCCGCCGCTTCAGACC-3’ 493/490
Gata6 5‘-ACCTTATGGCGTAGAAATGCTGAGGGTG-3’ 5‘-CTGAATACTTGAGGTCACTGTTCTCGGG-3’ 334/328
Brachyury 5‘-ATGCCAAAGAAAGAAACGAC-3’ 5‘-AGAGGCTGTAGAACATGATT-3’ 835
BMP2 5‘-GGGACCCGCTGTCTTCTAGT-3’ 5‘-TCAACTCAAATTCGCTGAGGAC-3’ 154
BMP4 5‘-ACTGCCGCAGCTTCTCTGAG-3’ 5‘-TTCTCCAGATGTTCTTCGTG-3’ 486
Osteocalcin 5’-CCTCTTGAAAGAGTGGGCTG-3 5’-CCTCGGGAGACAAACAACAT-3’ 268
Osteonectin 5’-GTCTCACTGGCTGTGTTGGA-3’ 5’-AAGACTTGCCATGTGGGTTC-3’ 264
Runx2 5’-GACGTTCCCAAGCATTTCAT-3’ 5’-ACTCTGGCTTTGGGAAGAG-3’ 181
Dspp 5'-GGAACTGCAGCACAGAATGA-3' 5'-CAGTGTTCCCCTGTTCGTTT-3' 199/193
Bcrp1 5'- GGTGAATCTCAGAACCATTGGGC-3’ 5'-GCTGTTGTCCGTTACATTGAATCC -3' 139
PPARγ 5’-CTGATGCACTGCCTATGAGC-3’ 5’-CAGACTCGGCACTCAATGGC-3’ 373
Hprt 5’-GTTAAGCAGTACAGCCCCAAA-3’ 5’-AGGGCATATCCAACAACAAACTT-3’ 131
Gapdh (Real-time) 5’-CCATCACCATCTTCCAGGAG-3’ 5’-GCATGGACTGTGGTCATGAG-3’ 322
(mouse/rat)