Supplemental Figure 1, Edwards et al. Proliferating Involuting · Matrigel ECFC HemSC HemSC+ECFC...
Transcript of Supplemental Figure 1, Edwards et al. Proliferating Involuting · Matrigel ECFC HemSC HemSC+ECFC...

Supplemental Figure 1. Proliferating and involutingIH morphology. Representative H&E staining ofproliferating (n=3) and involuting IHs (n=3). Boxed areaenlarged to right. Black arrowheads mark endothelialcells. Blue arrowheads mark perivascular cells. Blackarrows mark red blood cells. Scale bars - 100μm. IH,infantile hemangioma.
Prol
ifera
ting
Invo
lutin
gSupplemental Figure 1, Edwards et al.

Supplemental Figure 2, Edwards et al.
A αSMA MergedGLUT1
Prol
ifera
ting
Invo
lutin
g
Prol
ifera
ting
Prol
ifera
ting
Invo
lutin
g
C D NOTCH3 αSMA Merged
Prol
ifera
ting
Invo
lutin
g
CD31 MergedGLUT1
NOTCH3 CD31 Merged
Invo
lutin
g
Supplemental Figure 2. High magnification images of proliferating and involuting IHspresented in Figure 1. A) GLUT1 and CD31 co-staining. White arrowheads markGLUT1+/CD31+ cells. Yellow arrowheads mark GLUT1+/CD31- cells. 3, Proliferating IHn=2, involuting IH n=3. B) GLUT1 and αSMA co-staining. Yellow arrowheads markαSMA+/GLUT1- perivascular cells. Proliferating IH n=2, involuting IH n=7 C) NOTCH3 andCD31 co-staining. White arrowheads mark NOTCH3+/CD31+ cells. Yellow arrowheadsmark NOTCH3+/CD31- cells. Proliferating IH n=4, involuting IH n=5. D) NOTCH3 and αSMAco-staining. White arrowheads mark NOTCH3+/αSMA+ perivascular cells. Yellowarrowheads mark NOTCH3+/αSMA- lumenal cells. Proliferating IH n=5, involuting IH n=8.Scale bars - 25μm. Total number of IH specimens assessed for each antigen presented inSupplemental Table 3. αSMA, alpha smooth muscle actin; GLUT1, glucose transporter 1;IH, infantile hemangioma.
B

Supplemental Figure 3. NOTCH3 is expressed in asubset of Glut1+ IH cells. Proliferating and involuting IHspecimens stained for NOTCH3 and GLUT1. Whitearrowheads mark NOTCH3+/GLUT1+ cells. Yellowarrowheads mark GLUT1+/NOTCH3- lumenal cells.Proliferating IH n=4, involuting IH n=2 Scale bars - 50μm.GLUT1, glucose transporter 1; IH, infantile hemangioma.
Supplemental Figure 3, Edwards et al.
Prol
ifera
ting
Invo
lutin
g
Notch3 Glut1 Merged

Supplemental Figure 4, Edwards et al.
D CD31 MergedNG2
Prol
ifera
ting
Invo
lutin
g
B CD31 MergedPDGFRβ
Prol
ifera
ting
Invo
lutin
g
αSMA MergedPDGFRβPr
olife
ratin
gIn
volu
ting
A
C αSMA MergedNG2
Prol
ifera
ting
Invo
lutin
g
Supplemental Figure 4. High magnification images of proliferating and involuting IHspresented in Figure 2. A) PDGFRβ and αSMA co-staining. White arrowheads markPDGFRβ+/αSMA+ perivascular cells. Yellow arrowheads mark PDGFRβ+/αSMA- cells.Proliferating IH n=2, involuting IH n=3. B) PDGFRβ and CD31 co-staining. White arrowheadsmark PDGFRβ+/CD31+ cells. Yellow arrowheads mark PDGFRβ+/CD31- perivascular cells.Proliferating IH n=3, involuting IH n=2. C) NG2 and αSMA co-staining. White arrowheads markNG2+/αSMA+ cells, and yellow arrowheads mark NG2+/αSMA- cells. Proliferating IH n=7,involuting IH n=7. D) NG2 and CD31 co-staining. White arrowheads mark NG2+/CD31+ cells,and yellow arrowheads mark NG2+/CD31- cells. Proliferating IH n=7, involuting IH n=3. Scalebars - 25μm. Total number of IH specimens assessed for each antigen presented inSupplemental Table 3. αSMA, alpha smooth muscle actin; GLUT1, glucose transporter 1; IH,infantile hemangioma; NG2, neuron-glial antigen 2.

0
10
20
30
40
NOTCH3
PDGFRβ
NG2
αSMA
GLUT1
CD31
Supplemental Figure 5. Expression of endothelial and perivascular markers inisolated HemSCs. A) CD133+ HemSCs (n=3, done in duplicate) stained for NOTCH3,NG2, GLUT1, PDGFRβ, αSMA, and CD31. Scale bars - 50μm. B) NG2, GLUT1 andCD31 (red line) and control IgG (blue line) FACS of HemSCs (n=3, done in duplicate).C) Representative data of HemSC NOTCH3 and PDGFRB transcript levels determinedby qRT-PCR and normalized to BACTIN. (n=3, done in duplicate) Error bars represent ±S.D. αSMA, alpha smooth muscle actin; GLUT1, glucose transporter 1; IH, infantilehemangioma; HemSC, hemangioma stem cell; NG2, neuron-glial antigen 2.
A
B
Supplemental Figure 5, Edwards et al.
NOTCH3
Tran
scrip
t lev
els
(fg)
PDGFRB
NG2 GLUT1 CD31
C
0
10
20
30
40

Scr N3KD -0.5
0.0
0.5
1.0
1.5
Scr N3KD 0.0
0.5
1.0
1.5
2.0
Scr N3KD 0.0
0.5
1.0
1.5
Scr N3KD 0.0
0.5
1.0
1.5
Scr N3KD 0.0
0.5
1.0
1.5
Scr N3KD 0.0
0.5
1.0
1.5
n.s.
NOTCH1 HEY1
n.s.
HEY2
n.s.
Supplemental Figure 6. Characterization of NOTCH3knockdown HemSCs. A) Relative NOTCH1, HEY1 and HEY2transcript levels. Representative data from 4 independent N3KDHemSC populations and matching Scr HemSC populationsdetermined by qRT-PCR and normalized to BACTIN. Error barsrepresent ± S.D. n.s. - not significant. Student T-Test. B) RelativeVEGFA, ANG1, and ANG2 transcript levels. Representative datafrom 2 independent N3KD HemSC populations and matching ScrHemSC populations determined by qRT-PCR and normalized toBACTIN. Error bars represent ± S.D. *p<0.01, **p<0.005. Student T-Test. ANG1, angiopoietin 1; ANG2, angiopoietin 2; HemSC,hemangioma stem cell; N3KD, Notch3 knockdown,
Supplemental Figure 6, Edwards et al.
A
BVEGFA ANG1 ANG2
* ****
Scr N3KD0.0
0.5
1.0
1.5
Scr N3KD0.0
0.5
1.0
1.5
Scr N3KD0.0
0.5
1.0
1.5
Scr N3KD
0.0
0.5
1.0
1.5
Scr N3KD0.0
0.5
1.0
1.5
Scr N3KD0.0
0.5
1.0
1.5
Rel
ativ
eA
NG
1 le
vel
Rel
ativ
e V
EG
FAle
vel
Rel
ativ
e A
GN
2le
vel
2.0
Rel
ativ
e N
OTC
H1
leve
l
Rel
ativ
e H
EY
1le
vel
-0.05
Rel
ativ
e H
EY
2le
vel

Supplemental Figure 7, Edwards et al.
ScrH
emSC
N3K
D H
emSC
Growth mediaMural cell
differentiation media
Supplemental Figure 7. NOTCH3 knockdownblocks HemSC mural cell differentiation.Scrambled HemSCs (Scr HemSC) or NOTCH3knockdown HemSCs (N3KD HemSC) weregrown in growth or mural cell differentiationmedia for 2 weeks and stained for αSMA. Scalebars - 50μm. (n=3 HemSC populations done induplicate) αSMA, alpha smooth muscle actin;HemSC, hemangioma stem cell.
αSMA
αSMA αSMA
αSMA

Matrigel ECFC HemSC HemSC+ECFC
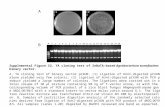
Supplemental Figure 8. HemSC/ECFC xenograftsrobustly develop an IH like phenotype. HemSCs,ECFCs or both ECFCs and HemSCs in 1:1 ratio wereresuspended in Matrigel, and subcutaneouslyimplanted into the flanks of immunocompromisedmice. Matrigel without cells served as a control.Xenografts were evaluated at Day 14 post-implantation. Two independent experiments were doneand representative data presented. Top) Grossappearance of explanted Matrigel. Bottom) H&Estaining of representative xenograft sections.Arrowheads mark large caliber red-blood cellcontaining IH-like vessel. Scale bar - 50μm. IH,infantile hemangioma; HemSCs, hemangioma stemcells; ECFCs, endothelial colony forming cells.
Supplemental Figure 8, Edwards et al.

Supplemental Figure 9. NOTCH3 knockdown in HemSCs inhibits IH development ina HemSC-only xenograft mouse model. A) Quantification of vessel density and caliber.Results representative of n = 3 HemSC populations (2 implants each). Error barsrepresent ± S.D. n.s., not significant, *p<0.0001. Student T-Test. B) αSMA+ mural celldensity determined as mean mural cell αSMA signal intensity normalized to IH endothelialGLUT1 signal intensity. Average mural cell αSMA expression determined as mean αSMAsignal intensity normalized DAPI+/αSMA+ cell number. Results representative of n = 3HemSC populations (2 implants each). Error bars represent ± S.D. n.s., not significant,**p<0.01. Student T-Test. αSMA, alpha smooth muscle actin; GLUT1, glucose transporter1; HemSC, hemangioma stem cell; MC, mural cell.
Supplemental Figure 9, Edwards et al.
n.s.
scr N3KD 0.0
0.1
0.2
0.3
0.4
0.5
Mea
n αS
MA
inte
nsity
Scr N3KD0.0
0.1
0.2
0.3
0.4
0.5
scr N3KD 0.0
0.5
1.0
1.5
2.0
2.5
**
0.0Scr N3KD
0.5
1.0
1.5
2.0
2.5
αSM
A+ M
C d
ensi
ty
scr N3KD 0
5
10
15
20
25
*
Scr N3KD0
5
10
15
20
25
Vess
el C
alib
er (μ
m)
scr N3KD 0.0
0.5
1.0
1.5
2.0 n.s.
Scr N3KD0.0
0.5
1.0
1.5
2.0Ve
ssel
den
sity
(#
ves
sels
/105
μm)
A B

A
SP EGF-like repeats 1-24 FC
0
0.5
1
1.5
2
2.5
3
Luci
fera
se F
old
Indu
ctio
n *" *"***
B
Human NOTCH3 decoy
JAG1
GSI
Ad-
+- + + +
+
- - -
- - - -
FC N3 decoy
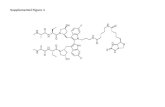
Supplemental Figure 10. The NOTCH3 inhibitor,human NOTCH3 Decoy (N3 Decoy), inhibits JAG1activation of Notch signaling. A) N3 Decoy encodes thesignal peptide (SP) and EGF-like repeats 1-24 of humanNOTCH3 fused to in frame with human IgG FC. B)Notch/CSL signal activation measured in HeLa cellsexpressing full-length rat NOTCH1, N3 Decoy and aNotch luciferase reporter (11CSL-Luc) co-cultured withHeLa cells expressing the Notch ligand, JAG1. Addition ofCompound E, a gamma-secretase inhibitor (GSI), wasused as a control and inhibited Notch activation of the11CSL-Luc reporter. Co-culture assays were performed intriplicate and repeated three times. Data presented asmean luciferase fold induction ± S.D. *p< 0.02, **p<0.005,Student T-Test. CSL, C promoter binding factor1/Suppressor of Hairless/Lag-1.
Supplemental Figure 10, Edwards et al.

A
FC Control NOTCH3 decoy
Day
21
B
Ad-N3 Decoy Ad-FcDay 0 Day 7 Day 21
HemSC/ECFCimplantation Harvest implant
US Doppler US Doppler
No injection
FC
C DControl Mice1 2 3 4
25 KD
1 2 3 4Fc 5N3 Decoy Mice
150 KD N3 decoy
Supplemental Figure 11. Design and validation of the NOTCH3 DecoyHemSC/ECFC xenograft study. A) Schematic of experimental time-line. B)Staining with antibodies against human FC of livers (Day 21) from mice injected anadenovirus encoding either N3 Decoy or FC. Liver sections from un-injected miceserved as a negative control. Scale bar - 50μm. C) Western blot with anti-FCantibody of sera (Day 21) collected from mice injected with the adenovirusencoding FC (n=4). D) Western blot of sera immunoprecipitated with antibodiesagainst human FC and then probed with anti-FC antibody (n=5). N3 Decoy was notdetected in the sera of mouse 4 and analysis of this xenograft was excluded fromthe study. ECFC, endothelial colony forming cell; HemSC, hemangioma stem cell;N3 Decoy; NOTCH3 Decoy
Supplemental Figure 11, Edwards et al.

CONTROL DECOY0
1
2
3
4
5
Supplemental Figure 12, Edwards et al.
αSMA αSMAFC NOTCH3 decoy
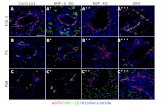
Supplemental Figure 12. NOTCH3 Decoy treatment has no effects on αSMA+perivasculature expression in hepatic vasculature. A) Liver sections from micetreated with FC (control) or NOTCH3 Decoy (n=2 populations; n=4 xenografts each)stained for αSMA. B) Mean αSMA expression determined as mean αSMA signalintensity normalized DAPI+/αSMA+ cell number. (n=2 populations; n=4 xenograftseach) n.s., not significant. Student T-Test. αSMA, alpha smooth muscle actin.
A B
n.s.
FC N3 Decoy
1
0
2
3
Mea
n αS
MA
inte
nsity 4
5