Src tyrosine kinase phosphorylation of nuclear receptor ...COS-7 cells cotransfected with HNF4α2...
Transcript of Src tyrosine kinase phosphorylation of nuclear receptor ...COS-7 cells cotransfected with HNF4α2...

Src tyrosine kinase phosphorylation of nuclearreceptor HNF4α correlates with isoform-specificloss of HNF4α in human colon cancerKarthikeyani Chellappaa, Lucy Jankovab, Jake M. Schnabla, Songqin Panc, Yann Brelivetd, Caroline L-S. Funge,Charles Chane, Owen F. Dentf, Stephen J. Clarkeg, Graham R. Robertsonb, and Frances M. Sladeka,1
aDepartment of Cell Biology and Neuroscience, University of California, Riverside, CA 92521; bCancer Pharmacology Unit, ANZAC Research Institute,University of Sydney, Sydney, New South Wales, Australia; c W. M. Keck Proteomics Laboratory, Institute for Integrative Genome Biology, University ofCalifornia, Riverside, CA 92521; dDépartement de Biologie et Génomique Structurales, Institut de Génétique et de Biologie Moléculaire et Cellulaire,67404 Illkirch, France; eDepartment of Anatomical Pathology, fDepartment of Colorectal Surgery, Concord Hospital; and gNorthern Clinical School,University of Sydney, Sydney, New South Wales, Australia
Edited by Tony Hunter, The Salk Institute for Biological Studies, La Jolla, CA, and approved December 15, 2011 (received for review April 28, 2011)
Src tyrosine kinase has long been implicated in colon cancer butmuch remains to be learned about its substrates. The nuclear re-ceptor hepatocyte nuclear factor 4α (HNF4α) has just recently beenimplicated in colon cancer but its role is poorly defined. Here weshow that c-Src phosphorylates human HNF4α on three tyrosinesin an interdependent and isoform-specific fashion. The initial phos-phorylation site is a Tyr residue (Y14) present in the N-terminal A/Bdomain of P1- but not P2-driven HNF4α. Phospho-Y14 interacts withthe Src SH2 domain, leading to the phosphorylation of two addi-tional tyrosines in the ligand binding domain (LBD) in P1-HNF4α.Phosphomimetic mutants in the LBD decrease P1-HNF4α proteinstability, nuclear localization and transactivation function. Immuno-histochemical analysis of approximately 450 human colon cancerspecimens (Stage III) reveals that P1-HNF4α is either lost or localizedin the cytoplasm in approximately 80% of tumors, and that stainingfor active Src correlates with those events in a subset of samples.Finally, three SNPs in the human HNF4α protein, two of whichare in the HNF4α F domain that interacts with the Src SH3 domain,increase phosphorylation by Src and decrease HNF4α protein stabi-lity and function, suggesting that individuals with those variantsmay be more susceptible to Src-mediated effects. This newly iden-tified interaction between Src kinase and HNF4α has importantimplications for colon and other cancers.
HNF4 isoforms ∣ SH2 SH3 domain ∣ SNP ∣ Src kinase ∣ tyrosinephosphorylation
Colon cancer, the third most common malignancy in the Uni-ted States, is a multifactorial disease that is influenced by
both genetics and the environment (1, 2). c-Src is a nonreceptortyrosine kinase that is strongly implicated in the development,growth, progression, and metastasis of several human cancers (3).In colon cancer, Src activation is associated with the early stages(4, 5) as well as progression and metastasis (6–8). Despite thislong association with colon cancer, much remains to be learnedabout Src substrates (9).
Hepatocyte nuclear factor 4alpha (HNF4α) (NR2A1) is ahighly conserved member of the nuclear receptor superfamilywith a recently identified endogenous ligand (linoleic acid) thatbinds in a reversible fashion (10, 11). HNF4α is best known for itsrole as a master regulator of liver-specific gene expression andas a key player in beta cells of the pancreas where it is mutatedin an inherited form of type 2 diabetes (12–14). HNF4α is alsoexpressed in kidney, stomach, and intestine; several recent papersalso show an important role for HNF4α in the colon (15–20).There are two different promoters (P1 and P2) of HNF4Athat are utilized in a temporal and tissue-specific fashion (11)(Fig. S1). While only P1-driven HNF4α (P1-HNF4α) is expressedin the adult liver, both P1- and P2-driven HNF4α (P2-HNF4α)are expressed in the adult intestine and colon (21, 22). Expression
of P1-HNF4α is decreased in several human cancers includinghepatocellular, gastric, renal, and colorectal carcinomas, whilethe expression of P2-HNF4α is either unchanged or upregulated(22, 23). However, the mechanism responsible for the differentialdysregulation of P1- and P2-HNF4α isoforms is not known. Herewe show that Src kinase preferentially phosphorylates P1-HNF4αin vitro and in vivo on multiple residues in a complex fashion,resulting in a loss of function and protein stability of P1- but notP2-HNF4α. We also show that the phosphorylation is influencedby the SH2 and SH3 domains of Src and by SNPs in HNF4α.Finally, we show that increased staining for active Src is asso-ciated with a loss of nuclear P1-HNF4α in a sizeable cohort ofhuman colorectal tumors. These findings suggest a unique linkbetween an oncogenic kinase, a potent differentiation factor andhuman colon cancer.
ResultsSrc Preferentially Phosphorylates P1-HNF4α Both In Vitro and In Vivo.An in vitro kinase assay showed that Src phosphorylates full lengthhuman P1-HNF4α2 as well as a truncated fragment that corre-sponds to the N-terminal portion [A/B and DNA binding domain(DBD)] (Fig. S2 A and B). Since P1- and P2-HNF4α differ byapproximately 29 amino acids in the A/B domain (Fig. S1C), werepeated the kinase assay and found that Src does not appreciablyphosphorylate P2-HNF4α8 in vitro (Fig. 1A and Fig. S2G).Cotransfection of HEK293 cells with constitutively active c-Src(c-Src Y530F) and HNF4α2 also showed in vivo tryosine phos-phorylation of P1-HNF4α2 using the phospho-Tyr (pY) specificAb 4G10; the signal was greatly reduced in cells pretreated withSrc inhibitor PP1 (Fig. S2H). In contrast, the in vivo tyrosine phos-phorylation signal of HNF4α8 was also much less than that ofHNF4α2, even in the presence of the tyrosine phosphatase inhibi-tor pervanadate (PV) (Fig. 1B).
To map Src phosphosites we mutated the two tyrosines in theHNF4α2 A/B domain to Phe (Y6F and Y14F) and found that onlytheY14Fmutation abolished the in vitro phosphorylation (Fig. S2C).Mass spectrometric analysis also identified a doubly phosphory-lated peptide containing Y277 and Y279 in the ligand bindingdomain (LBD) when P1-HNF4α2 was ectopically coexpressedwith c-Src Y530F in HEK293 cells (Fig. 1C); P2-HNF4α8 yielded
Author contributions: K.C., G.R.R., and F.M.S. designed research; K.C., L.J., S.P., and G.R.R.performed research; Y.B. performed computational modeling; K.C., J.M.S., and S.J.C.contributed new reagents/analytic tools; K.C., L.J., S.P., C.L.-S.F., C.C., O.F.D., G.R.R., andF.M.S. analyzed data; and K.C. and F.M.S. wrote the paper.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.1To whom correspondence should be addressed. E-mail: [email protected].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106799109/-/DCSupplemental.
2302–2307 ∣ PNAS ∣ February 14, 2012 ∣ vol. 109 ∣ no. 7 www.pnas.org/cgi/doi/10.1073/pnas.1106799109
Dow
nloa
ded
by g
uest
on
May
22,
202
1

6-fold less signal, despite equivalent recovery of the unphosphory-lated peptide (Fig. S2I). AY277/Y279 peptide containing a singlephospho group was also detected; sequencing of the peptide inthree independent experiments indicated that the phosphoryla-tion was on Y277, consistent with Y277 fitting the Src consensussite (DEEIYEEFF) better than Y279 (DNEY277AY279LK) (24).Y14 and Y277 were further validated as Src sites in in vivo and invitro assays using site-directed mutants and an Ab raised againstphospho-Y14 (pY14) (Figs. 1D and Fig. S2D and E). Importantly,the phosphorylation signal is almost completely lost in all Y14Fmutants, confirming that Y14 is critical for subsequent Src-mediated phosphorylation (Fig. 1D and Fig. S2F). Interestingly,a Y279F mutant consistently yielded increased total pYas well aspY14, but only in the presence of PV (Fig. 1D, and Fig. S2J). In-deed, in the absence of PV, the Y279F mutant resulted in reducedtotal pY and pY14 (Fig. S2J), suggesting a dynamic relationshipbetween Y279 and Y14 that may involve a phosphatase.
Phosphomimetic Mutants of P1-HNF4α Negatively Affect ProteinFunction and Stability. Modeling of the HNF4α LBD shows thatY277 flanks a salt bridge between E150 (helix 1) and K281 (helix8) (Fig. 2A, Top) that is part of a communication pathway linkingthe cofactor binding site to the dimer interface (25). In contrast,Y279 is near a hydrophobic cavity at the dimer interface, pointingtowards the interface of helix 9 and helix 1 (Fig. 2A, Bottom).Negatively charged groups at Y277 and Y279 are therefore pre-dicted to adversely affect HNF4α protein dimerization and/ortransactivation. The model also shows that Y277 and Y279 aretoo far apart (15.4 Å) for one kinase molecule to phosphorylateboth in a single event (Fig. S3A). This suggests that the phos-phorylation of these residues may occur in a sequential fashion,a notion that is supported by the singly phosphorylated Y277peptide detected by mass spectrometry.
To decipher the role of the three phosphorylated tyrosines,we examined the activity of single and double phosphomimeticmutants of Y14 (Y14E) and Y277/Y279 (Y277E/Y279E), respec-tively, and found that while Y14E had only a marginal effect onHNF4α2 transactivation (Fig. S3B), the Y277E/Y279E doublemutant completely abolished it (Fig. 2B). Since the DNA binding
activity of Y277E/Y279E was only marginally reduced comparedto that of WT HNF4α2 (Fig. S3C), we examined the intracellularlocalization of the double mutant and found a substantial cyto-plasmic localization (Fig. 2C, and Fig. S3D) that was reversedin the presence of nuclear export inhibitor leptomycin B (Fig. 2D).However, neither leptomycin B nor increased amounts of expres-sion vector (Fig. 2E), or addition of the potent HNF4α coactiva-tor PGC1α (Fig. 2F), recovered the transactivation function ofY277E/Y279E. Lastly, we examined the protein stability ofY277E/Y279E and found it greatly reduced compared to WTorY277F/Y279F (Fig. 2G and Fig. S3E). Taken together, theseresults indicate that Y277E/Y279E has reduced nuclear localiza-tion and protein stability. It also appears to have a reduced abilityto recruit coactivators and decreased transactivation function,consistent with active Src decreasing the ability of WT HNF4α2to activate transcription (Fig. 5E, and Fig. S5D and Fig. S6C).
Active Src Decreases P1- But Not P2-HNF4α Protein Stability In Vivo.c-Src Y530F also negatively affected HNF4α2 protein stability
A B
C D
Fig. 1. Src preferentially phosphorylates P1-HNF4α2 on Tyr14, Tyr 277, andTyr279. (A) In vitro Src kinase assay with HNF4α IP’d from bacterial lysatecontaining full-length GST-tagged P1- or P2-HNF4α (α2 and α8, respectively),recombinant Src kinase and 32P-γ-ATP. Shown is an autoradiograph of a blotfollowing SDS-PAGE. Bottom, IB with HNF4α Ab. (B) In vivo Src kinase assay ofCOS-7 cells cotransfected with HNF4α2 and c-Src WT or Y530F and treated100 μM PV for different time points as indicated. IP’d HNF4α was probedwith anti-pY (4G10) or HNF4α Ab. (C) Mass spectrometry (nano-LC/MS/MS)analysis of HNF4α IP’d from HEK293 cells cotransfected with c-Src Y530F andHNF4α2 or HNF4α8 after a 10-min PV treatment. A peptide containing Y277/Y279 was detected at m∕z 1471.2, +2, corresponding to a doubly phosphory-lated fragment. The identity of the 1471.2, +2 peptide was verified using asynthetic peptide corresponding to the peptide sequence shown. (D) In vivoSrc kinase assay as in (B) but with WT or Y to F mutants of HNF4α2 cotrans-fected with c-Src Y530F into HEK293 cells and treated with 100 μM PV for10 min prior to lysis. pY14, phospho-specific Ab to pY14 HNF4α.
Fig. 2. Tyr277 and Tyr279 are key residues for Src-mediated effects onHNF4α. (A) Three-dimensional model of human HNF4α LBD in the region ofY277 and Y279. The two subunits of the homodimer are shown in cyan andgreen. Key residues and helices are indicated. (B) Luciferase activity (relativelight units, RLU) from transiently transfected Cos-7 cells with WT or doublemutants of HNF4α2 and an HNF4α-responsive luciferase construct (ApoB.-85-47.E4.Luc). Data are the means of triplicate samples from one experiment;error bars show s.d. *P < 0.002 WT vs. Y277E/Y279E mutant. (C) Subcellularlocalization of WT and double mutants of Y277 and Y279 in HNF4α2 ex-pressed in COS-7 cells. Immunolabeled cells (α445 Ab) were visualized usinga Zeiss 510 confocal microscope (40×) and digitally magnified. (D) Subcellularlocalization as in (C) but after leptomycin B (LMB) or vehicle treatment for8 h of HNF4α2 Y277E/ Y279E mutant. Immunolabeled cells (α445 Ab) werevisualized with a Nikon Eclipse Ti inverted microscope (20×) and digitallymagnified. (E) Fold difference (normalized RLU in the presence or absenceof HNF4α2) of COS-7 cells transfected with WT or Y277E/Y279E mutant ofHNF4α2 and ApoAI-4.Luc reporter treated with LMB or vehicle for 9 h. Dataare the means of triplicate samples from one experiment; error bars shows.d. *P < 0.05 vehicle vs. LMB for WT, n.s., no significant change. (F) Folddifference (normalized RLU in the presence or absence of PGC1α) of WT orY277E/Y279E mutant of HNF4α2 cotransfected in HEK293 cells with ApoAI-4.Luc reporter and CMV.β-gal. *P < 0.0005 fold difference in mock-trans-fected vs. WT HNF4α2. (G) Quantification of protein stability of WT anddouble mutants of Y277 and Y279 in HNF4α2 expressed in Cos-7 cells in thepresence of 50 μMCHX at the indicated time points. Experiments in B–Gwereperformed two to three times with similar results.
Chellappa et al. PNAS ∣ February 14, 2012 ∣ vol. 109 ∣ no. 7 ∣ 2303
BIOCH
EMISTR
Y
Dow
nloa
ded
by g
uest
on
May
22,
202
1

in an HEK293 cotransfection assay, while Src inhibitor PP2increased HNF4α2 protein levels (Fig. 3A and Fig. S4A, Right).Proteasome inhibitors (MG132 and ALLN) (Fig. 4A, Right andFig. S4A, Left) substantially increased HNF4α2 protein levels, butlysosomal inhibitors (chloroquine and ammonium chloride) hadonly a marginal effect (Fig. S4A), suggesting that Src decreasesHNF4α2 protein stability mainly via the proteasome pathway. Toexamine the effect of endogenous Src activation on endogenousHNF4α, we treated human colorectal cancer cells CaCo2, whichexpress both P1- and P2-HNF4α isoforms, with EGF to induceSrc activity (Figs. S4 B and C). The results show a loss ofP1- but not P2-HNF4α upon EGF treatment at later time points(Figs. 3B and Fig. S4D). We also show that endogenous P1- butnot P2-HNF4α is phosphorylated in CaCo2 cells by both IP/IB(Fig. 3C) and mass spectrometry; the latter showed that as muchas 40% of the P1-HNF4α was doubly phosphorylated at Y277/Y279 in CaCo2 cells (Fig. 3D). Finally, a 24-h treatment ofCaCo2 cells with Src inhibitor PP2 significantly decreased endo-genous P1-HNF4α phosphorylation (Fig. 3E) and increased thelevel of P1-HNF4α protein (Fig. 3F). In contrast, there was amarked decrease in P2-HNF4α protein upon PP2 treatment,which could be due to the fact that P1-HNF4α is known to de-crease transcription of the P2 promoter (21). All told, selectivephosphorylation of endogenous P1-HNF4α in a colon cancer cellline correlates with decreased protein stability and is consistentwith the experiments using ectopic expression and phosphomi-metic mutants.
Src SH2 and SH3 Domains Interact with P1-HNF4α. Since Src SH2 andSH3 domains (bind pYand proline-rich regions, respectively) areknown to play a role in regulating Src activity and processivephosphorylation of its substrates, we examined their ability tointeract with HNF4α. Not only did WT HNF4α2 phosphorylatedby active Src bind the Src SH2 domain in vitro, but Y14 wasrequired for maximal interaction (Fig. 4A, and Fig. S5B). In con-trast, P2-HNF4α8 did not bind Src SH2 appreciably (Fig. 4B). Toverify the specificity of the interaction, we showed that the SrcSH2 mutant R178A does not interact with phospho-HNF4α2(Fig. S5B), and that the pY14 peptide binds directly to the WTSrc SH2 domain, but not the R178A mutant (Fig. 4C). We alsofound that the pY14 but not the nonphosphorylated (Y14) pep-
tide can effectively compete with phospho-HNF4α2 for bindingthe Src SH2 domain (Fig. 4D). Finally, using an in vivo kinaseassay, we show that a WT Src SH2 domain is required for totaltyrosine phosphorylation of HNF4α2 but is less important forY14 phosphorylation (Fig. 4E). These results are consistent withthe notion that phosphorylation of Y14 is the first event and thatthe Src SH2 domain subsequently binds pY14. The results are
P1-HNF4
PP2: - + - +
24 h 48 h
P2-HNF41.0 2.4 1.0 2.09
1.0 0.14 1.0 0.24
CaCo2
100
%
0
1391.2
Y277/Y279
1471.2
pY277/pY279
pY277/pY279
Y277/Y279= 0.41
0
50
100
150
200
250
CHX (h): 0.5 1 2 4 6 9
HN
F4
: EG
F v
s U
ntre
ated
(%
)
P1-HNF4P2-HNF4
CaCo2: EGF B
Vehicle MG132
HNF4 2
-actin 1.0 4.6
P1-HNF4
pY14
IP: rIgG HNF4
Input mIgG 4G10
IP
P1-HNF4
P2-HNF4
C
D
IP
IP: 4G10mIgG
3 h 24 hStart Start - + - +
Input NormalizedPhospho P1-HNF4 1.0 1.1 1.0 0.41
PP2P1-HNF4 :
E F
A HEK 293: HNF4 2 + c-Src Y530F
Rel
ativ
e H
NF
42
(%)
CHX (h): 0.75 1.5 3 60
20
40
60
80
100
120
140
**
(-) c-Src Y530F
(+) c-Src Y530F
0
20
40
60
80
100
120
140
0
50
100
150
200
250
Fig. 3. Active Src decreases P1- but not P2-HNF4α protein stability; tyrosine phosphorylation of endogenous P1-HNF4α. (A) Left, Protein stability ofWT HNF4α2expressed in HEK293 cells in the presence or absence of c-Src Y530F. Graph represents HNF4α protein level normalized to Coomassie stain. Data are means oftriplicates from one of at least 10 representative experiments; error bars show s.e.m. *P < 0.05.Right top, Right, IB of HNF4α2 and β-actin from whole cellextracts of HEK293 treated with either DMSO or 50 μM MG132 for 8 h. Shown are representative blots from three independent experiments. (B) Proteinstability of P1- and P2-HNF4α in CaCo2 cells harvested at the indicated time points after EGF + CHX treatment. Shown is one representative quantificationof EGF-treated versus untreated controls from five independent experiments. (C) In vivo phosphorylation of endogenous HNF4α from CaCo2 cells. Top, IP’dphospho-Tyr proteins (4G10) were IB’d with either P1- or P2-HNF4α Abs. Bottom, IP’d HNF4α (α445) was probed with pY14 or P1-HNF4α Ab. (D) Mass spectro-metry analysis as in Fig. 1C but of P1-HNF4α IP’d (αN1.14 Ab) from CaCo2 cells. Signals corresponding to the nonphosphorylated Y277/Y279 (m∕z 1391.2, +2) andpY277pY279 (m∕z 1471.2, +2) peptides were detected. Ratio of the amount of pY277/pY279 to Y277/Y279 is indicated. (E) As in (C, Top) but from CaCo2 cellstreated with ethanol or 10 μMPP2 for different time points. Values corresponding to normalized phospho P1-HNF4α (IP’d P1-HNF4α divided by input P1-HNF4α)are shown. (F) IB of P1-HNF4α and P2-HNF4α from whole cell extracts of CaCo2 cells after 24 h or 48 h treatment with 10 μM PP2 or DMSO. Shown are triplicatesfrom one of three independent experiments.
A B
C D E
Fig. 4. Src SH2 domain interacts with pY14 and is critical for phosphorylationof Y277 and Y279. (A) Pulldown assay with GST or GST.Src-SH2 proteins andwhole cell extracts from HEK293 cells cotransfected with c-Src Y530F andWT or the indicated Y to F mutants of HNF4α2. IBs with α445 show theamount of HNF4α in the Input and after the Pulldown. (B) As in (A) but withhuman HNF4α2 and HNF4α8. (C) Pulldown assays as in (A) but with WT orR178A mutants of Src SH2 and 5 μg of pY14 peptide (Fig. S2D). Shown is anIB of bound peptide after Pulldown spotted onto PVDF membrane and de-tected by an Ab that recognizes the pY14 peptide. (D) Pulldown as in (A)but the interaction between WT HNF4α2 and GST.Src-SH2 was competedwith either the pY14 or the Y14 peptide. Shown is the IB with the HNF4αantibody from one representative experiment and quantification of boundHNF4α relative to no peptide from 5 and 2 independent experiments forpY14 and Y14 competition, respectively. Error bar represents s.e.m. *P < 0.01relative to no peptide competition. (E) In vivo Src kinase assay in HEK293 cellscotransfected in 100-mm plates with HNF4α2WT (5 μg) and either c-Src Y530For Src SH2 mutant (c-Src R178A/Y530F) at different ratios of transfectedDNA (5 μg to 0.5 μg). IP’d HNF4αwas probed with 4G10, pY14 or P1/P2 HNF4αAb. Indicated is the fold difference in the phosphorylation signal with theSrc SH2 mutant relative to c-Src Y530F at each ratio. Fold difference forthe 1∶0.2 ratio was calculated using a darker exposure of the 4G10 blot. NA,not applicable as no signal was observed.
2304 ∣ www.pnas.org/cgi/doi/10.1073/pnas.1106799109 Chellappa et al.
Dow
nloa
ded
by g
uest
on
May
22,
202
1

also of interest since Y14 (PApY14TTL) only partially fits theknown consensus for a Src kinase substrate (pY(E/D/T)(E/N/Y)(L/I) ) and SH2 domain ligand [(P/H)(I/P)pY(E/D/V/I)(L/I/E)(I/L/V)] (24). Finally, to show that pY14 is required for phosphor-ylation of the LBD, we examined an N-terminally truncatedHNF4α2 construct (ΔAB) and found that it was neither phos-phorylated in vivo by active Src (Fig. S6A) nor greatly repressedin transactivation function by Src compared to WT HNF4α2(Fig. S6C). These results confirm that the P1-driven A/B domainis actively required for the phosphorylation of the LBD.
Since the F domain of HNF4α is proline rich, we predicted andobserved an interaction between WT HNF4α2 and the Src SH3domain in a GST pulldown assay (Fig. 5A). Interestingly, like theSH2 mutant, an SH3 mutant W121A decreased total HNF4α2phosphorylation but had little effect on pY14 (Fig. 5B). Theseresults suggest that the Src SH3 domain, like the SH2 domain,is not required for Y14 phosphorylation but that it is required forphosphorylation of the LBD. Consistent with this notion, massspectrometry did not show any phosphorylation on Y277 whenHNF4α2 was cotransfected with the Src SH3 mutant (Fig. S5C).Finally, a luciferase assay showed that while both the WT andc-Src Y530F decreased HNF4α2 transcriptional activity, the SH3mutant W121A and, to a lesser extent the SH2 mutant R178A,exhibited less repression (Fig. S5D). Taken together, these resultsshow that HNF4α2 binds both the Src SH2 and SH3 domains andthat those interactions play an important role in the isoform-specific phosphorylation of HNF4α.
SNPs in HNF4α Affect Src Phosphorylation and Protein Stability.Thereare three SNPs in human HNF4α that might be relevant tothe interaction with Src (dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/). Introduction of mutations corresponding totwo SNPs in the F domain (P421L, rs6031602, and P436S,rs1063239) dramatically decreased binding to the Src SH3 do-
main (Fig. 5A). Rather unexpectedly, in the presence of cotrans-fected active Src, both variants (P421L and P436S) yielded agreater pY14 signal compared to WT HNF4α2 (Fig. 5C), espe-cially P421L. Another SNP adjacent to Y279 (L280F, rs6093980)also yielded a greater pY14 signal (Fig. 5C). Importantly, all threeSNPs (L280F, P421L, P436S) exhibited decreased protein stabi-lity compared to WT HNF4α2, but only in the presence of activeSrc (Fig. 5D and Fig. S6D). Finally, two of the SNP variants(L280F and P421L) exhibited lower levels of transactivation thanWT HNF4α2 in the absence of active Src, while the third variantP436S activated transcription as well as WT (Fig. 5E). Impor-tantly, all three variants were more susceptible to Src-mediatedrepression of transactivation than WT HNF4α2, even after nor-malizing to the amount of HNF4α protein (Fig. 5E). This in-creased sensitivity of the SNP variants to Src is consistent with theincreased phosphorylation observed in Fig. 5C.
The results thus far show that the proline-rich F domain ofWT HNF4α2 interacts with the Src SH3 domain and that this in-teraction is required for full phosphorylation. However, the SNPvariants show that the WT F domain may inhibit Src-mediatedphosphorylation and repression of HNF4α2 transactivation. Toaddress this apparent discrepancy, we examined the in vivophosphorylation of HNF4α2 truncated in the F domain (ΔF). Theresults show that HNF4α ΔF is indeed phosphorylated by Src(Fig. S6A). In addition, mass spectrometry of cotransfectedHNF4α and active Src showed that the amount of the singly phos-phorylated pY277 peptide (1431.2) was greater in ΔF than in WTHNF4α2 (41% vs 3.6%, respectively) (Fig. S6B). Consistent withthe notion that the F domain inhibits Src-mediated phosphoryla-tion of HNF4α2, the ΔF truncation also exhibited a greater re-duction in transactivation in the presence of active Src than didWTHNF4α2 (Fig. S6C). Therefore, the F domain appears to playa modulatory role in Src-mediated effects on HNF4α2 via bindingthe SH3 domain.
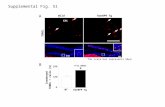
Preferential Loss of Nuclear P1-HNF4α Correlates with Active SrcStaining in Human Colon Cancer. In a pilot study of tumors fromhuman patients with colon cancer, we found that only 26 out of43 tumors were positive for a P1-HNF4α specific Ab (P1) whileall 43 tumors showed staining with the Ab that recognizes bothP1- and P2-HNF4α (P1/P2) (Fig. S7 A and B). This preferentialloss of P1-HNF4α is consistent with previous results on a similar-sized cohort of patients (22, 23). Screening a larger cohort of 450consecutive Stage III colon cancer cases revealed four categoriesof tumors when intensity of P1 staining was considered—loss ofall P1 staining (a, approximately 12%); predominantly cytoplas-mic P1 staining (b, approximately 18–24%); nuclear and cytoplas-mic P1 staining (c, approximately 47–49%); and only nuclearP1 staining (d, approximately 18–23%) (Figs. 6 A–B, Fig. S7 Cand D). All told, approximately 80% of tumor samples showeda P1-HNF4α staining that was not exclusively nuclear; in contrast,in normal tissue HNF4α staining was observed only in the nucleus(Figs. S7E and S8B).
To assess the state of Src activation in the tumors, a subset ofthe samples (98 tissue microarray cores from invasive front andcentral regions of tumors) were stained with anti-phospho-Srcantibody (pTyr 419, clone 9A6). The tumors showed a consistentstaining for pSrc in the nucleus, especially when P1-HNF4α waseither completely cytoplasmic or absent (Fig. 6C, sections I, II). Incontrast, when there was appreciable nuclear P1-HNF4α in thetumor area, there was low pSrc staining (sections III, IV). Further-more, the adjacent normal tissue, which had essentially exclusivenuclear P1-HNF4α in nearly all the cells, showed little evidenceof pSrc staining (Fig. S8C). All told, the majority of the samples(Fig. S8C, categories 1 + 2, 61 out of 98 tumor cores; X2 ¼ 7.772,p ¼ 0.005) were consistent with the notion that activation of Srcmay result in cytoplasmic localization and/or complete loss ofP1-HNF4α. Here we report that cytoplasmic P1-HNF4α staining
3,000
2,000
1,000
2,000
1,000
A
B
E
D
C
Fig. 5. Src SH3 domain interacts with proline-rich F domain of HNF4α; SNPsin HNF4α2 LBD and F domain affect Src-mediated phosphorylation. (A) Pull-down assays with GST.Src-SH3 protein and whole cell extract from HEK293cells transfected with WTor the indicated SNP variants of HNF4α2. (B) In vivoSrc kinase assay of HEK293 cells cotransfected with HNF4α2 and either c-SrcY530F or Src SH3 mutant (c-Src W121A/Y530F). IP’d HNF4α was probed with4G10, pY14, or P1/P2 HNF4α Ab. (C) In vivo Src kinase assay as in Fig. 1B butwith HNF4α2WTor the indicated SNP variants cotransfected with c-Src Y530Finto HEK293 cells. Right, quantification of the pY14 signal relative to the to-tal HNF4α signal from one of three representative experiments. (D) Proteinstability of WT and SNP variants of HNF4α2 cotransfected with c-Src Y530Finto HEK293 cells. Shown is the average of triplicates from one of two inde-pendent experiments of the HNF4α IB signal normalized to Coomassie stain.*P < 0.02 SNP variants vs. WT. (E) Transcriptional activity ofWTor SNP variantsof HNF4α2 either in the presence or absence of c-Src Y530F in HEK293 cellscotransfected with pZL.HIV.LTR.AI-4 reporter and CMV.βgal. Shown is theRLU normalized to HNF4α protein and β-gal activity for WT (Left) and % ofWT (set to 100%) for SNP variants (Right). Data are the meanþ ∕ − s:e:m: oftwo independent experiments performed in triplicates. *P < 0.05 for SNPvariants in the presence vs. the absence of c-Src Y530F.
Chellappa et al. PNAS ∣ February 14, 2012 ∣ vol. 109 ∣ no. 7 ∣ 2305
BIOCH
EMISTR
Y
Dow
nloa
ded
by g
uest
on
May
22,
202
1

in human colon cancer correlates with staining for active Src.While active Src is typically found in the cytoplasm and is teth-ered to the plasma membrane via a fatty acid anchor, there is aprevious report of nuclear localization of pSrc in human coloncancer (26). We have also observed nuclear pSrc in CaCo2 cells,as well as in three other human colon cancer cell lines (RKO,SW620, SW480) (Fig. S8D).
DiscussionIn this study, we identify HNF4α as a new target of Src kinase andin the process elucidate a complex interaction that has importantramifications for human colon cancer. We find that Src firstphosphorylates Y14 in the N-terminal A/B domain of P1-HNF4α(Fig. 6D, Step 1). Subsequently, pY14 binds the SH2 domain ofSrc and the proline-rich F domain of HNF4α binds the Src SH3domain, thereby facilitating the phosphorylation of first Y277 andthen Y279 in the HNF4α LBD (Step 2). We propose that thistethering of the Src protein brings its kinase domain in closeproximity to the Y277/279 residues that are normally not veryaccessible. Since we have shown previously that the HNF4α Fdomain functions as a transcriptional repressor in part by inter-acting with the LBD (27), we also propose that by binding the Fdomain, the Src SH3 domain disrupts the interaction with theLBD, thereby making Y277 and Y279 more accessible for phos-phorylation. The net result is a triply phosphorylated HNF4α(Step 3) in which pY277 and pY279 evidently disrupt the coacti-vator binding site, resulting in a loss of transactivation, nuclearlocalization, and protein stability. We also show that three SNPsin HNF4α (L280F, P421L, and P436S) increase Src-mediatedphosphorylation, degradation of HNF4α protein and repressionof transcription. Therefore, we propose that while the SNP var-iants in the F domain disrupt the interaction with the Src SH3domain, they also abrogate the interaction of the F domain withthe LBD, thereby at least partially exposing Y277 and Y279(Fig. 6E). What is not clear, however, is exactly how the SNPslead to increased phosphorylation of Y14. Importantly, this entiresequence of events happens only on P1-HNF4α as P2-HNF4αdoes not contain Y14. Since P2-HNF4α8 has the same F domainas P1-HNF4α2, this (and other experiments presented in thiswork) indicates that the binding of the Src SH3 domain to HNF4αalone is not sufficient to cause substantial phosphorylation of
HNF4α. As we observed in the human colon cancer samples, thenet result is a preferential loss of P1- but not P2-HNF4α underconditions that activate c-Src, or a Src kinase family member(the pSrc antibody recognizes the pY419 of several different Srcfamily kinase members). Furthermore, there appears to be anegative effect of Src on HNF4α-mediated transactivation, evenwhen reduced HNF4α protein levels are taken into account. Thissuggests that any phospho-HNF4α protein remaining in thenucleus may also have reduced transcriptional activity.
While a few other nuclear receptors are known to be phos-phorylated by Src kinase (28–30), here we report the isoform-specific phosphorylation of a nuclear receptor by Src kinase. Wealso provide evidence suggesting that SNP variants in a nuclearreceptor may increase one’s susceptibility to c-Src, and hence toSrc-mediated cancer. Finally, the interaction between Src kinaseand HNF4α described here may also be relevant in other cancers,such as hepatocellular carcinoma, which has been shown to havehigh levels of active Src (31) and a loss of P1-driven HNF4α (22).
Materials and MethodsSrc Kinase Assays. In vitro kinase assays were performed as per the manufac-turer’s protocol (Millipore) using recombinant active Src (25–50 ng), 32P-γ-ATPand purified GST fusion proteins or human HNF4α [wild type (WT) or mutant]immunoprecipitated (IP’d) from transfected HEK293 cells. In vivo kinaseassays used HEK293 or COS-7 cells transfected with HNF4α and c-Src Y530Ftreated with 100 μM PV for 3–30 min before lysing with RIPA buffer asindicated. HNF4α was IP’d with the affinity purified Ab α445 and then elutedwith 445 peptide. In vivo phosphorylation assays in CaCo2 cells were per-formed with cells at <50% confluency; whole cell extracts were IP’d witheither α445 Ab (and peptide eluted) or 4G10 platinum Ab (Millipore). Allreaction products were analyzed by 10% SDS-PAGE followed by autoradio-graphy or immunoblot (IB) analysis. All Src kinase assays were performedthree or more times.
Human Colonic Adenocarcinoma Tissue Staining. Patients had a surgical resec-tion for Australian Clinicopathological Stage III colonic adenocarcinoma,equivalent to TNM Stage III (32, 33) but were excluded if they had a historyof ulcerative colitis (UC) or Crohn’s disease or if UC, Crohn’s disease, or poly-posis coli was detected in the surgical specimen. Tissue microarrays (TMAs)were constructed with 1.0 mm cores from morphologically representativeareas of the original archived paraffin blocks (the central region of the tumor,deep advancing tumor front, and adjacent nonneoplastic mucosa) using anAdvanced Tissue Arrayer, ATA-100 (Chemicon). HNF4α immunoreactivity in
Fig. 6. Preferential loss and cytoplasmic staining of P1-HNF4α in human colon cancer correlates with active Src; model of multistep phosphorylation of HNF4αby Src. (A) Diverse patterns of nuclear and cytoplasmic accumulation of HNF4α protein as detected by the P1-specific HNF4αAb (brown staining) in colon cancersfrom different patients: a) total absence of P1-HNF4α in malignant epithelial cells; b) predominantly cytoplasmic with many nuclei negative for P1-HNF4α; c)cytoplasmic and nuclear P1-HNF4α; and d) strong nuclear with almost no cytoplasmic presence of P1-HNF4α. Magnification 200×. (B) Number of tumors with agiven staining pattern of P1-HNF4α (a, b, c or d) shown in (A) out of the total cohort of 405. (C) Representative stains for P1-HNF4α and active Src (pSrc) in asubset of tumors: P1-HNF4α (I, III); pSrc (II, IV). (D, E) Model of multistep phosphorylation of P1-HNF4α by Src tyrosine kinase and effect of SNP’s in the HNF4α2LBD (L280F) and F domain (P421L, P436S).
2306 ∣ www.pnas.org/cgi/doi/10.1073/pnas.1106799109 Chellappa et al.
Dow
nloa
ded
by g
uest
on
May
22,
202
1

each tissue core was assessed independently by L.J. and two experiencedpathologists (C.L-S.F., C.C.) for staining intensity (0 ¼ absent, 1 ¼ weak,2 ¼ intermediate, 3 ¼ strong staining) using P1-specific and P1/P2 antibodiesas previously described (34). pSrc (pTyr419) immunoreactivity was similarlydetermined. Approval for this study incorporating informed consent wasobtained from the South Western Sydney Area Health Service.
ACKNOWLEDGMENTS. We thank P. Chapuis and L. Bokey for establishing thetumor bank at Concord Hospital; C. Clarke for TMA construction; D. Morasand B. Fang HNF4α LBD and SH2 structure analysis; and C. Wu for GST.Src.SH3.The clinical component was supported by a Cancer Institute New SouthWalesTranslational Program Grant for Colorectal Cancer. All other work wasfunded by an National Institutes of Health R01 grant to F.M.S. (DK053892).
1. Walther A, et al. (2009) Genetic prognostic and predictive markers in colorectal cancer.Nat Rev Cancer 9:489–499.
2. ACS (2008) Colorectal Cancer Facts & Figures 2008-2010 (American Cancer Society,Atlanta).
3. Yeatman TJ (2004) A renaissance for SRC. Nat Rev Cancer 4:470–480.4. Iravani S, et al. (1998) Elevated c-Src protein expression is an early event in colonic
neoplasia. Lab Invest 78:365–371.5. Cartwright CA, Meisler AI, Eckhart W (1990) Activation of the pp60c-src protein kinase
is an early event in colonic carcinogenesis. Proc Natl Acad Sci USA 87:558–562.6. Summy JM, Gallick GE (2003) Src family kinases in tumor progression and metastasis.
Cancer Metastasis Rev 22:337–358.7. Talamonti MS, Roh MS, Curley SA, Gallick GE (1993) Increase in activity and level of
pp60c-src in progressive stages of human colorectal cancer. J Clin Invest 91:53–60.8. Termuhlen PM, Curley SA, Talamonti MS, Saboorian MH, Gallick GE (1993) Site-specific
differences in pp60c-src activity in human colorectal metastases. J Surg Res 54:293–298.9. Amanchy R, et al. (2008) Identification of c-Src tyrosine kinase substrates using mass
spectrometry and peptide microarrays. J Proteome Res 7:3900–3910.10. Yuan X, et al. (2009) Identification of an endogenous ligand bound to a native orphan
nuclear receptor. PLoS One 4:e5609.11. Bolotin E, Schnabl J, Sladek F (2009) HNF4A (Homo sapiens)., Transcription Factor
Encyclopedia, http://www.cisreg.ca/cgi-bin/tfe/home.pl.12. Bolotin E, et al. (2010) Integrated approach for the identification of human hepato-
cyte nuclear factor 4alpha target genes using protein bindingmicroarrays.Hepatology51:642–653.
13. Odom DT, et al. (2004) Control of pancreas and liver gene expression by HNF transcrip-tion factors. Science 303:1378–1381.
14. Gupta RK, Kaestner KH (2004) HNF-4alpha: from MODY to late-onset type 2 diabetes.Trends Mol Med 10:521–524.
15. Garrison WD, et al. (2006) Hepatocyte nuclear factor 4alpha is essential for embryonicdevelopment of the mouse colon. Gastroenterology 130:1207–1220.
16. Darsigny M, et al. (2009) Loss of hepatocyte-nuclear-factor-4alpha affects colonic iontransport and causes chronic inflammation resembling inflammatory bowel disease inmice. PLoS One 4:e7609.
17. Babeu JP, Darsigny M, Lussier CR, Boudreau F (2009) Hepatocyte nuclear factor 4alphacontributes to an intestinal epithelial phenotype in vitro and plays a partial role inmouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol297:G124–134.
18. Ahn SH, et al. (2008) Hepatocyte nuclear factor 4alpha in the intestinal epithelial cellsprotects against inflammatory bowel disease. Inflamm Bowel Dis 14:908–920.
19. Darsigny M, et al. (2010) Hepatocyte nuclear factor-4alpha promotes gut neoplasia inmice and protects against the production of reactive oxygen species. Cancer Res70:9423–9433.
20. Cattin AL, et al. (2009) Hepatocyte nuclear factor 4alpha, a key factor for homeostasis,cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol29:6294–6308.
21. Briancon N, et al. (2004) Expression of the alpha7 isoform of hepatocyte nuclear factor(HNF) 4 is activated by HNF6/OC-2 and HNF1 and repressed by HNF4alpha1 in the liver.J Biol Chem 279:33398–33408.
22. Tanaka T, et al. (2006) Dysregulated expression of P1 and P2 promoter-drivenhepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol208:662–672.
23. Oshima T, et al. (2007) Downregulated P1 promoter-driven hepatocyte nuclear factor-4alpha expression in human colorectal carcinoma is a new prognostic factor againstliver metastasis. Pathol Int 57:82–90.
24. Huang H, et al. (2008) Defining the specificity space of the human SRC homology 2domain. Mol Cell Proteomics 7:768–784.
25. Brelivet Y, Kammerer S, Rochel N, Poch O, Moras D (2004) Signature of the oligomericbehaviour of nuclear receptors at the sequence and structural level. EMBO Rep5:423–429.
26. Sakai T, Kawakatsu H, Fujita M, Yano J, OwadaMK (1998) An epitope localized in c-Srcnegative regulatory domain is a potential marker in early stage of colonic neoplasms.Lab Invest 78:219–225.
27. Sladek FM, Ruse MD, Jr, Nepomuceno L, Huang SM, Stallcup MR (1999) Modulationof transcriptional activation and coactivator interaction by a splicing variation inthe F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol19:6509–6522.
28. Kim H, Laing M, Muller W (2005) c-Src-null mice exhibit defects in normal mammarygland development and ERalpha signaling. Oncogene 24:5629–5636.
29. Buitrago C, Vazquez G, De Boland AR, Boland RL (2000) Activation of Src kinase inskeletal muscle cells by 1, 1,25-(OH(2))-vitamin D(3) correlates with tyrosine phos-phorylation of the vitamin D receptor (VDR) and VDR-Src interaction. J Cell Biochem79:274–281.
30. Mahajan NP, et al. (2007) Activated Cdc42-associated kinase Ack1 promotes prostatecancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad SciUSA 104:8438–8443.
31. Masaki T, et al. (1998) pp60c-src activation in hepatocellular carcinoma of humans andLEC rats. Hepatology 27:1257–1264.
32. Davis NC, Newland RC (1983) The reporting of colorectal cancer: The AustralianClinico-pathological Staging (ACPS) System. Med J Aust 1:282.
33. Fielding LP, et al. (1991) Clinicopathological staging for colorectal cancer: An Interna-tional Documentation System (IDS) and an International Comprehensive AnatomicalTerminology (ICAT). J Gastroenterol Hepatol 6:325–344.
34. Kishimoto T, et al. (2008) A case of alpha-fetoprotein-producing pulmonary carcinomawith restricted expression of hepatocyte nuclear factor-4alpha in hepatoid foci: A casereport with studies of previous cases. Hum Pathol 39:1115–1120.
Chellappa et al. PNAS ∣ February 14, 2012 ∣ vol. 109 ∣ no. 7 ∣ 2307
BIOCH
EMISTR
Y
Dow
nloa
ded
by g
uest
on
May
22,
202
1