Non-covalent protein-ligand interactions? Easy as Pi
-
Upload
david-thompson -
Category
Education
-
view
1.112 -
download
2
description
Transcript of Non-covalent protein-ligand interactions? Easy as Pi

N l i li d i i ? Non-covalent protein-ligand interactions? Easy as π
Jonathan Lai David C. ThompsonMing Hong-Hao Ingo MüggeMing Hong-Hao Ingo Mügge

π hides a wealth of complexity
It is the ratio of circumference (c) to diameter (d)
It is an irrational number
c
d =dc
It is an irrational number
It is a transcendental number 3. 1415926535 8979323846 26433832795028841971 6939937510 58209749445923078164 0628620899 86280348
d
It occurs in lots of interesting places:
• Normalization of the normal distribution
445923078164 0628620899 86280348253421170679 8214808651 32823066470938446095 5058223172 53594081284811174502 8410270193 85211055596446229489 5493038196 44288109
• Distribution of the primes
• Buffon’s needle problem
756659334461 2847564823 37867831652712019091 4564856692 34603486104543266482 1339360726 02491412737245870066 0631558817 48815209209628292540 9171536436 78925903• The Bible (I Kings 7:23 and Chronicles 4:2)209628292540 9171536436 78925903600113305305 4882046652 13841469519415116094 3305727036 57595919530921861173 8193261179 31051185480744623799 6274956735 18857527π is tricky. So are π-π molecular interactions
31 August 20112
248912279381 8301194912 . . .π is tricky. So are π π molecular interactions

Presentation Overview: The What, Why, and How of it all
What are we trying to do?
Why are we trying to do it?
Perform a detailed investigation of π- π stacking interactions between heterocyclic ring systems and aromatic amino acid side-chains
Why are we trying to do it?
Could we preferentially suggest heterocycles for synthesis?Can we extract any generalities about geometry?
How are we going to do it?
Can we extract any generalities about geometry?
Extract deposited structural information from the PDBEvaluate interactions using ab initio methodologies
3

Why are π-π interactions important?
Prevalent dispersive driven interactions
St t ll d ti ll i t t i t d i t l l Structurally and energetically important inter- and intra-molecular interactions[1]
Sandwich T-shaped Parallel Displaced
1 33 k l/ l[2] 2 24 k l/ l[2] 2 22 k l/ l[2]
4
-1.33 kcal/mol[2] -2.24 kcal/mol[2] -2.22 kcal/mol[2]
[1] A Medicinal Chemist’s Guide to Molecular Interactions, Caterina Bissantz, Bernd Kuhn, Martin Stahl JMC, Article ASAP (2010)[2] M. O. Sinnokrot, and C. David Sherrill, J. Phys. Chem. A, 110, 10656-10668 (2006) [CCSD(T) / aug-cc-pVDZ (carbon) / cc-pVDZ (hydrogen)]

Why are π-π interactions important? Viramune® (nevirapine)
NH2
CHC
HO
ODistance: 4.43Å
Tilt Angle: 16.88°
E
Nevirapine
H2C
ΔIEEst.: -1.38 kcal/mol
Nevirapine
1VRT.pdbOHTyrosine
HN
O
NN N
11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2',3'-e]
5
6H dipyrido[3,2 b:2 ,3 e][1,4]diazepin-6-one

Relibase+[3] Search Query
Filtering Criteria:
ki i i
Focused on compounds of these motifs:
• π-π stacking interactions
• Crystals w/ resolutions: ≤ 2.5Å
NN N
N
N
N N
N
• Not nucleic acids
• No duplicates by PDB ID Ignored substituents
N
• Aromatic, planar ligands Excluded compounds of these motifs:
O
N
O
Radius: 3.4Å ≤ x ≤ 4.1Å
Tilt angle: 0 ≤ x ≤ 40°
Geometric Filtering criteria:
6
O
NRing pucker: |x| ≤ 5°
[3] Relibase+, v2.2

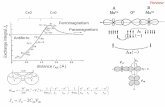
Heteroatom Count ( # of Ring systems found in data set)
Motif Histidine Phenylalanine Tyrosine Tryptophan
136 114 93 94136 114 93 94
45 67 165 8245 67 165 82
98 307 212 10198 307 212 101
P i (1 4 0 11 4 4Pyrazine (1,4 diazine), Triazine,
and Others
0 11 4 4
7
Radius: 3.4Å ≤ x ≤ 4.1Å
Tilt angle: 0 ≤ x ≤ 40°
Ring pucker: |x| ≤ 5°

Non-covalently Bound Set (Histidine)
Two distinct clusters
1. “T-shaped”1. T shaped
2. “Sandwich/Displaced sandwich”
Conformations between clusters are still accessible (e g radius 4 5Å tilt angle 30°)
p
accessible (e.g. radius ~ 4.5Å, tilt angle ~ 30 )
Can still extract a wealth of information (e g Can still extract a wealth of information (e.g. ligand orientation, minimum contact distance)
8
Radius: 3.0Å ≤ x ≤ 5.0Å
Tilt angle: 0 ≤ x ≤ 80°
Ring pucker: |x| ≤ 5°

Non-covalently bound Amino Acid Set
HIS PHE
9
TRPTYRRadius: 3.0Å ≤ x ≤ 5.0Å
Tilt angle: 0 ≤ x ≤ 80°
Ring pucker: |x| ≤ 5°

How did we use MOE?
Custom MOE browser
• Review complexes from PDB• Review complexes from PDB
• Automatically prepare Q-CHEM[4] input
— CCSD(T) / aug-cc-pVDZ basis
— Account for BSSE (EAB, EA, and Eb)
• Triage to ensure correct bond orders
10
• Interface with HPC environment for job submission
[4] Q-CHEM, v3.2

What do the results tell us?
A little more than we could possibly understand
11
Shape: Position with respect to plane of His. (Below) (Above) Size & Colour: CCSD(T) Energy (kcal/mol) [Small and red is a very attractive interaction]

Density Functional Theory (DFT) and Dispersion
DFT is formally exact in the ground state, ifthe exact form of EXC[ρ] is known
• It is not
DFT migrated from solid-state physics intothe chemistry community to great effecty y g
• Local density approximation works wellfor materials, not so well for molecules
D
• Add non-locality to better approximateinhomogeneity of molecular density– Perdew’s ‘Jacob’s Ladder of functionals’
There are a number of well-known issues involving DFT, including its inability to dealwith dispersion[5]with dispersion[5]
[5] C. David Sherrill, J. Chem. Phys., 132, 110902 (2010) 12

Density Functional Theory (DFT) and Dispersion
1
CCSD(T) / kcal/mol
-1
0
-4.5 -3.5 -2.5 -1.5 -0.5 0.5 1.5
CCSD(T) / kcal/mol
R² = 0.941
-2
D[6
]/
kcal
/mol
R 0.941
-4
-3
ωB9
7X-D
6
-5
-6[6] Jeng-Da Chai and Martin Head-Gordon, Phys. Chem. Chem. Phys. 10, 6615 (2008)
13

Dig into the data[7]
Above (A) Below (C)Below (B)
Above (45)
Volumetricl t iclustering
Below (B)
Below (52)
Below (C)• Focus on pyrimidine (98 structures)
• Use volumetric clustering
14
Correlation with overall dipole?[7] boyd, danah. 2010. "Privacy and Publicity in the Context of Big Data." WWW. Raleigh, North Carolina, April 29

AM1 Dipole Moment versus ωB97X-D for pyrimidinecontaining systems
0
1 2 3 4 5 6
AM1 Dipole Moment
-2
-1
-3
-2
X-D
kca
l / m
ol
R² = 0.6402-4ω
B97X
6
-5
-6
15
Electrostatic component masks dispersive interaction, making our quest for acanonical geometry somewhat redundant

Outlook and Future Work
Robust workflow in place to extract π-π interactions and torun QM calculations within our HPC environment
i i d b di l d d i h• π-π interactions tend to be displaced sandwiches• Potential energy surface is shallow• Calculating and analyzing interaction potential energy surfaceg y g p gy
is challenging• ωB97X-D appears to be a viable alternative to CCSD(T)• Substantial electrostatic component to interaction energy• Substantial electrostatic component to interaction energy
— Correlation of pyrimidine energy with total AM1 dipolemoment
Select suitable, representative, displaced sandwich geometryand evaluate heterocyclic systems
16
and evaluate heterocyclic systems

Cultural Highlight
Ethnographic examination of ‘financiers’financiers
— Investment bankers
Risk managers— Risk managers
— Fund managers
“All models are wrong, but some models are useful” – G. E. P. Boxmodels are useful G. E. P. Box
“If tit d i l i it i b tt t b “If exactitude is elusive, it is better to be approximately right than certifiably wrong” – B. B. Mandelbrot
The Big Short, Michael Lewis, W. W. Norton (2010)

Acknowledgements
Dr. Sandy Farmer
Dr. Miguel TeodoroDr. Miguel Teodoro
Dr. Bryan McKibben
18