log iBAQ control IP
Transcript of log iBAQ control IP

shFBW7
shControl
Actin
USP9X
FBW7α
A
Flag-FBW7β
shUSP9xMG132
+ + + +
− + − +shControl
USP9X
Flag-FBW7β
Actin
B
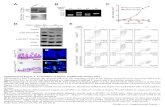
Supplementary Figure 1. (A) iBAQ plot showing MS-enrichment of FBW7 and interaction partners over a control IgG. (B) Western blots for indicated proteins in cells transfected with indicated shRNAs. (C) Western blots for indicated proteins in cells co-transfected with Flag-FBW7β and indicated shRNAs.
46
810
log
iBAQ
FBW
7 IP
4 6 8 10log iBAQ control IP
FBW7USP9X C

Flag-c-Jun + +
+ + HA-Ub
GST-USP9X − +
Ubi
-c-J
UN
WB: FlagIP: HA
260
140
100
75
50
40
HA-Ubiquitin & Flag-FBW7overexpression in HCT116
IP: HA/HA-peptideElution
Flag-M2 IP on pooled sequentially eluted HA-ubiquitin complexes
3x wash
Ubi-Crest Assay on beads bound HA-ubiquitylated FBW7
Western blotting for K48 specific linkage
A B
No DUB
USP2OTU
BAIN1
AMSHUSP9X
Ubi
-Fla
g-FB
W7α
α−K48: WB
α-Flag: WB
UM
K48
-Ubi
-FB
W7α
Stripped/reprobed
Supplementary Figure 2. USP9X cleaves K48-linked polyubiquitin chains on FBW7. (A) Recombinant USP9X does not affect polyubiquitination of c-JUN in an in vitro deubiquitylation reaction. (B) Schematic for experiment in C. (C) Ubiquitin chain restriction (UbiCRest) experiment with indicated deubiquitinases: USP2 (promiscuous), OTUBAIN1 (K48-linked), and AMSH (K63-linked).
C
Flag INPUT
USP9X

shU9 #3
shControl
shU9 #4
USP9x
c-Jun
c-Myc
Actin
HCT116
Cyclin E
Supplementary Figure 3. Accumulation of SCF(FBW7) substrates in cells transfected with USP9X-shRNAcompared to a non-targeting control.

A B
Olfm4 ISH
Usp
9xΔ
GU
sp9x
+/+
MCM6
Alkaline phosphatase
C D
Usp
9xΔ
GU
sp9x
+/+
Usp
9xΔ
GU
sp9x
+/+
Usp
9xΔ
GU
sp9x
+/+
Chromogranin
E
0
1.0
2
0.5
Usp9xFbw7
1.5
Rel
ativ
e m
RN
A ex
pres
sion
(
norm
aliz
ed to
Act
in)
Usp9x+/+
Usp9xΔG
*
Usp
9xΔ
GU
sp9x
+/+
BrdU
0
3
6
9
Usp9x+/+
Usp9xΔG
Brd
U+
cells
/col
onic
cry
pt
*
F
Supplementary Figure 4. (A) IHC sections stained for proliferating cells (MCM6) from the intestine of indicated mice. Scale bar = 50 μm. (B) BrdU staining for proliferating cells in colonic crypts from indicated mice, quantification shown in right panel. Scale bar = 100 μm, n = 3–4 mice/group. (C) In situ hybridization for Olfm4 (stem cells) on the sections from the intestine of indicated mice. Scale bar = 100 μm. (D and E) IHC for enterocytes (Alkaline phosphatase) and enteroendocrine cells (Chromogranin) in gut from indicated mice. Scale bars = 100 μm. (F) qRT-PCR analysis showing mRNA levels of indicated genes normalised to actin and represented as fold change over control, in isolated crypts from Usp9x+/+ (wildtype) and Usp9xΔG gut, n = 5–8 mice/group.

0
2
4
6G
oble
t cel
ls/v
illus
c-MycΔG/+
c-JunΔG/+
Usp9xΔG
Usp9x
+/+0
2
4
6
8
10
12
Brd
U+
cells
/cry
pt
Vehicle DBZ
Usp9xΔG
A B
-
Supplementary Figure 5. (A) Average goblet cell number per villus from indicated mice. (B) Average BrdU+ cell number per crypt from indicated mice.

A B170
150
130
110
90
70
501 2 3 4 5 6 7 8 9 10 110
(weeks)
Per
cent
wei
ght
Usp9x+/+
Usp9xΔG
p <0.0001
1 2 3 4 5 6 7 8 9 10 110(weeks)
150
130
110
90
70
50
Fbw7+/+
Fbw7ΔG/+
p <0.05Per
cent
wei
ght
Supplementary Figure 6. (A and B) Weight curves presented as percent of starting weight in indicated mice from colitis-driven tumorigenesis experiment, n = 5–9 animals/genotype. P values were calculated by Log-rank (Mantel-Cox) test.

SupplementaryTable.ShortlistedcandidatesfromIP-massspectrometryexperimentusingendogenousFBW7asbait.
GeneSymbol Description FunctionFBXW7 F-box/WDrepeat-containingprotein7 E3LigaseSKP1 S-phasekinase-associatedprotein1 SCFComponentUBAP2L Ubiquitin-associatedprotein2-like UbiquitylationMYCBP2 MYCbindingprotein2 ProbableE3ubiquitin-proteinligaseHERC2 E3ubiquitin-proteinligaseHERC2 E3LigaseHUWE1 E3ubiquitin-proteinligaseHUWE1 E3LigaseHECTD1 E3ubiquitin-proteinligaseHECTD1 E3LigaseUSP9X Probableubiquitincarboxyl-terminalhydrolaseFAF-X DeubiquitinaseUSP20 Ubiquitincarboxyl-terminalhydrolase20 DeubiquitinaseFKBP8 Peptidyl-prolylcis-transisomeraseFKBP8 ChaperonFKBP4 Peptidyl-prolylcis-transisomeraseFKBP4 ChaperonPSMA2 Proteasomesubunitalphatype-2 ProteasomesubunitPSMA3 Proteasomesubunitalphatype-3 ProteasomesubunitPSMA6 Proteasomesubunitalphatype ProteasomesubunitPSMD3 26Sproteasomenon-ATPaseregulatorysubunit3 ProteasomesubunitFOXP2 ForkheadboxproteinP2 Transcriptionfactorc-JUN TranscriptionfactorAP-1 TranscriptionfactorSTAT1 Signaltransducerandactivatoroftranscription1-alpha/beta TranscriptionfactorMSH6 DNAmismatchrepairproteinMsh6 DNAreplication/GenomeintegrityMCM3 DNAreplicationlicensingfactorMCM3 DNAreplication/GenomeintegrityMCM7 DNAreplicationlicensingfactorMCM7 DNAreplication/GenomeintegrityTOP1 DNAtopoisomerase1 DNAreplication/GenomeintegrityRAD21 Double-strand-breakrepairproteinrad21homolog DNAreplication/GenomeintegrityNAP1L1 Nucleosomeassemblyprotein1-like1 DNAreplication/GenomeintegrityHDAC2 Histonedeacetylase2;Histonedeacetylase DNAreplication/GenomeintegrityCHD4 Chromodomain-helicase-DNA-bindingprotein4 DNAreplication/GenomeintegrityMRE11A Double-strandbreakrepairproteinMRE11A DNAreplication/GenomeintegrityDNAJB6 DnaJhomologsubfamilyBmember6 DNAreplication/GenomeintegrityMTOR Serine/threonine-proteinkinasemTOR KinaseSTK3 Serine/threonine-proteinkinase3 KinasePANK2 Pantothenatekinase2,mitochondrial KinaseGALK1 Galactokinase KinaseDEK ProteinDEK KinaseCSNK1A1 CaseinkinaseIisoformalpha KinasePI4KB Phosphatidylinositol4-kinasebeta KinaseWEE1 Wee1-likeproteinkinase KinaseXPO1 Exportin-1 NuclearexportCSE1L Exportin-2 NuclearexportKPNB1 Importinsubunitbeta-1 NuclearimportRANBP1 Ran-specificGTPase-activatingprotein TransportLUC7L PutativeRNA-bindingproteinLuc7-like1 RNAbinding/processing

DDX39 ATP-dependentRNAhelicaseDDX39A RNAbinding/processingMARS Methionine-tRNAligase,cytoplasmic RNAbinding/processingRAE1 mRNAexportfactor RNAbinding/processingBCAS2 Pre-mRNA-splicingfactorSPF27 RNAbinding/processingLARS Leucine--tRNAligase,cytoplasmic RNAbinding/processingRBM39 RNA-bindingprotein39 RNAbinding/processingRBM28 RNA-bindingprotein28 RNAbinding/processingDDX24 ATP-dependentRNAhelicaseDDX24 RNAbinding/processingTARS Threonine--tRNAligase,cytoplasmic RNAbinding/processingFMR1 FragileXmentalretardationprotein1 RNAbinding/processingIARS Isoleucine--tRNAligase,cytoplasmic RNAbinding/processingYBX2 DNA-bindingproteinA;Y-box-bindingprotein2 RNAbinding/processingMLL Histone-lysineN-methyltransferase EnzymeRPN2 DPDglycosyltransferasesubunit2 EnzymeDPM1 Dolichol-phosphatemannosyltransferase EnzymePRMT1 ProteinarginineN-methyltransferase1 EnzymeACLY ATP-citratesynthase EnzymeEIF4G1 Eukaryotictranslationinitiationfactor4gamma1 TranslationEIF3F Eukaryotictranslationinitiationfactor3subunitF TranslationEIF2B4 TranslationinitiationfactoreIF-2Bsubunitdelta TranslationEIF5B Eukaryotictranslationinitiationfactor5B TranslationMFF Mitochondrialfissionfactor MitochondrialSSBP1 Single-strandedDNA-bindingprotein,mitochondrial MitochondrialATP5J2 ATPsynthasesubunitf,mitochondrial MitochondrialMTCH2 Mitochondrialcarrierhomolog2 MitochondrialMAGED2 Melanoma-associatedantigenD2 CelladhesionACTR2 Actin-relatedprotein2 CytoskeletonTUBA1C Tubulinalpha-1Cchain CytoskeletonTUBB2A Tubulinbeta-2Achain Cytoskeleton