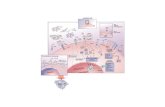
Electron Transport Chain/Respiratory Chain Proton gradient formed Four large protein complexes...
-
Upload
richard-medina -
Category
Documents
-
view
215 -
download
0
Transcript of Electron Transport Chain/Respiratory Chain Proton gradient formed Four large protein complexes...

Electron Transport Chain/Respiratory Chain
Proton gradient formed
Four large protein complexes
Mitochondria localized Energetically favorable electron flow

Mitochondrion Inner Membrane
Respiration site
Surface area for humans ca. 3 football fields
Highly impermeable (no mitochondrial porins)
Matrix and cytoplasmic sides

Standard Reduction Potentials

ΔG˚΄ = -nF Δ E˚΄ F = 96,480 J mol-1 V-1
Favorable Electron Flow: NADH to O2
Net electron flow through electron transport chain:
½O2 + 2H+ + 2e- H2O ΔE˚΄ = + 0.82V
NAD+ + H+ + 2e- NADH ΔE˚΄ = - 0.32V
Subtracting reaction B from A:
½O2 + NADH + H+ H2O + NAD+ ΔE˚΄ = + 1.14V
ΔG˚΄ = -220 kJ mol-1

Electron Transport Energetic’s

Electron Transport Chain Components
Protein complexes:
I.NADH-Q reductase
II.Succinate dehydrogenase
III.Cytochrome C reductase
IV.Cytochrome C oxidase
Bridging components:
Coenzyme Q and Cytochrome C What is the driving force for this electron flow?

Coupled Electron-Proton Transfer Through NADH-Q Oxidoreductase
FMN bridges: NADH 2 e- donor with FeS 1 e- acceptor
L-shaped Complex I
Overall reaction:
NADH + Q + 5H+ NAD+ + QH2 + 4H+

Coupled Electron-Proton Transfer Through NADH-Q Oxidoreductase
H+ movement with 1 NADHIron-sulfur clusters (a.k.a.
nonheme-iron proteins)
2Fe – 2S or 4Fe – 4S complexes

NADH-Q Oxidoreductase (Complex I) Structure
Largest of respiratory complexes
Mammalian system contains 45 polypeptide subunits; 8 Fe-S complexes; 60transmembranehelices

Different Quinone (Q) Oxidation States
QH2 generated by complex I & II
Membrane-bound bridging molecule
Overall reaction:
QH2 + 2Cyt Cox + 2H+ Q + 2Cyt Cred + 4H+
X

Oxaloacetate Enzyme Regeneration from Succinate
• Succinate Dehydrogenase
• Fumerase
• Malate Dehydrogenase

Pathways that Contribute to the Ubiquinol Pool Without Utilizing Complex I

Alternative Q-Cycle Entry PointsComplex I
Complex II (citric acid cycle)
Glycerol 3-phosphate shuttle
Fatty acid oxidation (electron-transferring-flavoprotein dehydrogenase)

Electron-Transport Chain Reactions in the Mitochondria

The Q CycleElectron transfer to Cytochrome c Reductase via 3 hemes and a Rieske iron-sulfur center Overall reaction:
QH2 + 2Cyt Cox + 2H+
Q + 2Cyt Cred + 4H+
ISP – iron sulfur protein

The Q Cycle

Cytochrome c Oxidoreductase StructureIntermembrane sideHeme-containing homodimer
with 11 subunit monomers
Functions to:
• Transfer e- to Cyt c
• Pump protons into the intermembrane space
Matrix side

Cytochrome c Oxidase: Proton Pumping and O2 Reduction

Cytochrome c Oxidase: O2 Reduction to H2OReaction shown:2Cyt Cred + 2H+ + ½ O2
2Cyt Cox + H2O
Overall reaction:
2Cyt Cred + 4H+ + ½ O2
2Cyt Cox + H2O + 2H+

Cytochrome c Oxidase
O2 to H2O reduction site
Intermembrane space
Matrix
Oxygen requiring step
13 subunits; 10 encoded by nuclear DNA
CuA/CuA prosthetic group positioned near intermembrane space

Cytochrome c Oxidase

Electron-Transport Chain Reactions in the Mitochondria

Electron-Transport Chain Reactions in the Mitochondria

Mitochondrial Electron-Transport Chain Components

ATP Synthesis via a Proton Gradient
The two major 20th century biological discoveries:
DNA structure andATP synthesis

ATP-Driven Rotation in ATP-Synthase: Direct Observation
γ rotation with ATP present
With low ATP 120-degreeIncremental rotation
Glass microscope slide

ATP Synthase with a Proton-Conducting (F0) and Catalytic (F1) Unit
Matrix side
Intermembrane side
F1 matrix unit contains 5 polypeptide chain types (α3, β3, γ, δ, ε)
Proton flow from intermembrane space to matrix

Matrix side
ATP-Synthase with Non-Equivalent Nucleotide Binding Sites
Side view
F1 contains:
α3, β3 heximeric ring and γ, ε central stalk
Central stalk andC-ring form therotor andremainingmolecule is the stator
Top view

γ-Rotation Induces a Conformational Shift in the β Subunits
Each β subunit interacts differently with the γ subunit
ATP hydrolysis can rotate the γ subunit

Proton Flow Around C-Ring Powers ATP SynthesisSubunit C Asp protonation favors movement out of hydrophylic Subunit a to membrane region
Deprotonation favors Subunit a movement back in contact with Subunit a

Proton Motion Across the Membrane Drives C-Ring Rotation

C-Ring Tightly Linked to γ and ε Subunits
C-ring rotation causes the γ and ε subunits to turn inside the α3β3 hexamer unit of F1
Columnar subunits (2 b) with δ prevent rotation of the α3β3 hexamer unit
What is the proton to ATP generation ratio?

Mitochondrial ATP-ADP Translocase
Net movement down the concentration gradient for ATP (out of matrix) and ADP (into matrix)
No energy cost

Mitochondrial Transporters for ATP Synthesis
Net movement against the concentration gradient for Pi (into matrix) and charge balance -OH (out of matrix)
Proton gradient energy cost

ATP Yield With Complete Glucose Oxidation

Heat Generation by an Uncoupling Protein UCP-1Brown adipose tissue rich in mitochondria for heat generation
Pigs nest, shiver, and have large litters to compensate for lack of brown fat

ATP Synthesis Chemical Uncoupling
What physiological effect might DNP have in humans?

Electron Transport Chain Inhibitors
Toxins (e.g. fish and rodent poison rotenone)
Site specific inhibition for biochemical studies
What impact will rotenone have on respiration (O2 consumption)?

Proton Gradient Central to Biological Power Transmission

Problems: 13, 21, 23, 31, 33