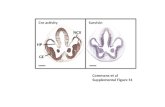
Cre f/f Normal trol Cre · A B Supplemental Figure 1.Generation of ARNT knockout mice. (A) Breeding...
Transcript of Cre f/f Normal trol Cre · A B Supplemental Figure 1.Generation of ARNT knockout mice. (A) Breeding...

A B
Supplemental Figure 1. Generation of ARNT knockout mice. (A) Breeding of ARNTf/f (denoted as f/f) mice with αMHC-MCM
(denoted as Cre) mice to generate MCM/ARNTf/f (denoted as Cre, f/f) mice. (B) Protocol for oral administration of tamoxifen in
different experimental groups. Cardiac function was assessed at 2 and 4 weeks after the completion of tamoxifen treatment.
Arrows refer to the time when echocardiography was performed. (C) Mean body weight and food intake in controls, αMHC-
MCM (represented as Cre) and ARNTf/f (represented as f/f) and αMHC-MCM/ARNTf/f (represented as cre, f/f) mice treated
with normal and tamoxifen chow (n=7-11 independent experiments). Data are presented as mean ± SEM. * P < 0.01 versus
normal chow.
4W
Cre, f/f
Cre
f/f
Cre, f/f
Co
ntr
ol
csA
RN
T-/
-
Tamoxifen
0 2W
Tamoxifen
Tamoxifen
Normal
chow
Cre f/f
f/f
f/f
x
f/+ Cre, f/+
Cre,f/+
x
f/+Cre,f/fB
od
y w
eig
ht
(g)
0
5
10
15
20
25
Cre f/f Cre/ff Cre f/f Cre/ff
NS NS
Normal chow Tamoxifen chow
C
Cre, f/f Cre, f/f

0
0.2
0.4
0.6
0.8
1
1.2
1.4 ControlARNT-/-
05000
10000150002000025000300003500040000
ControlARNT-/-
*
AR
NT
mR
NA
leve
ls
(rel
ativ
e ex
pres
sion
)
A
C
Supplemental Figure 2. ARNT expression in the heart and other organs of control and csARNT-/- mice. (A) ARNT mRNA levels(as determined by RT-PCR) of cardiac samples from control and csARNT-/- mice after tamoxifen administration at different timepoints (n=6 independent experiments). (B) ARNT mRNA levels in different organs of control and csARNT-/- mice (n=6independent experiments). (C) Western blot analysis of ARNT in different organs of control and csARNT-/- mice 4 weeks aftertamoxifen treatment. Data are presented as mean ± SEM. * P < 0.01.
Normal chowTamoxifen chow
AR
NT
mR
NA
leve
ls(fo
ld c
hang
e)
B
Adipose Brain Kidney Liver Lung Spleen
ARNT
GAPDH
*
** ** *
csARNT-/-
csARNT-/-

Normal chowTamoxifen
Normal chowNormal chow
010203040506070
05
101520253035
%EF %FS
Normal chowTamoxifen
0
0.2
0.4
0.6
0.8
IVS-
d
0
1
2
3
4
5
TamoxifenTamoxifen
LVID
-d (m
m)
NS NS
NS NS
M-m
ode
αMHC-MCM, ARNTf/f
Tamoxifen chow Normal chow A
B C
D E
Supplemental Figure 3. Cardiac function in three sets of control animals by echocardiographic assessment. (A) representativeM-mode images from indicated mice at 12 weeks. EF (B), FS (C), LVID-d (D), and interventricular septal thickness at end-diastole (IVS-d) (E) (n=10-12 independent experiments). Data are presented as mean ± SEM.
αMHC-MCM ARNTf/f
ARNTf/f Cre,ARNTf/f
Cre ARNTf/f Cre,ARNTf/f
Cre
ARNTf/f Cre,ARNTf/f
Cre ARNTf/f Cre,ARNTf/f
Cre

Supplemental Figure 4. Morphologic changes of control (ARNTf/f littermates) and csARNT-/- mice 4 weeks after tamoxifentreatment. (A) Ratio of heart weight to tibia length (n=12 independent experiments). (B) Ratio of lung weight to tibia length(n=12 independent experiments). (C) Tibia length (n=12 independent experiments). Data are presented as mean ± SEM. * P< 0.01.
012345678 *
Hea
rt w
eigh
t/tib
ia le
ngth
(m
g/m
m)
0
2
4
6
8
10
12
0
5
10
15*
NS
Lung
wei
ght/t
ibia
leng
th
(mg/
mm
)
Tibi
a le
ngth
(mm
)
A B C
Control csARNT-/- Control csARNT-/- Control csARNT-/-

0
100
200
300
400
500
600
700
800
900
Control ARNT-/-
0
1000
2000
3000
4000
5000
6000
7000
8000
Control ARNT-/-
AN
P m
RN
A
BN
P m
RN
A
**
csARNT-/- csARNT-/-
Supplemental Figure 5. mRNA levels of (A) atrial natriuretic peptide (ANP) and (B) brain natriuretic peptide (BNP) in the
hearts of control and csARNT-/- mice at 4 weeks after tamoxifen administration (n=6 independent experiments). Data are
presented as mean ± SEM. * P < 0.01.
A B

*
TAG
con
tent
(nm
ol/µ
g pr
otei
n)0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
Control siRNA ARNT siRNA
ARNT
Tubulin
Control siRNA ARNT siRNA
00.20.40.60.8
11.2
AR
NT
expr
essi
on
(fold
cha
nge)
ControlsiRNA
ARNTsiRNA
*
*
B C
0
50
100
150
200
250
300
350
400
control csARNT-/-
TAG
(mg/
dl)
NSA
Supplemental Figure 6. Serum triacylglycerol (TAG) level with ARNT knockdown. (A) Serum TAG levels in csARNT-/- hearts(n=7 independent experiments). (B) Western blot analysis of ARNT expression in NRCM treated with ARNT siRNA and controlsiRNA for 48 hours ( n=3 independent experiments). (C) TAG levels in NRCM treated with control or ARNT siRNA for 48 hours(n=6 independent experiments). Data are presented as mean ± SEM. * P < 0.01.
csARNT-/-

D
Supplemental Figure 7. Assessment of myocardial glucose metabolism in vivo and in vitro. (A) Glucose oxidation measuredin working hearts (n=6 independent experiments). (B) mRNA levels of HKII, Glut1 and Glut4 determined by qRT-PCR oncardiac muscle samples of control and csARNT-/- mice (n=5-6 mice per genotype). (C) Glucose uptake as measured by 14C-labeled glucose in NRCM after treatment with control and ARNT siRNA (n=6 independent experiments). (D) Extracellularacidification rate (ECAR) measured with the XF24 Extracellular Flux Analyzer in NRCM transfected with control or ARNT siRNA(n= 6 independent experiments). Data are shown as mean ± SEM. * P < 0.01 vs control.
Glu
cose
oxi
datio
n (n
mol
/g d
ry w
t/min
)
BA
0
5
10
15
20
25
30
ControlsiRNA
ARNTsiRNA
ECA
Rm
PH/m
in/m
g pr
otei
n
*m
RN
A le
vels
(fold
cha
nge)
0
0.2
0.4
0.6
0.8
1
1.2
1.4 Control
csARNT-/-
* *
0100200300400500600700
ControlsiRNA
ARNTsiRNA
*
Glu
cose
upt
ake
(cpm
/µg
prot
ein)
C
0
200
400
600
800
1000
1200
1400
1600
Control csARNT-/-
*
Glut1 Glut4HKII
*

00.5
11.5
22.5
3
ControlsiRNA
ARNTsiRNA
PPARα
Control siRNA ARNT siRNA
Tubulin
Rel
ativ
e in
tens
ity(fo
ld c
hang
e)
*
Supplemental Figure 8. Expression of PPARα in NRCM treated with control and ARNT siRNA for 48 hours. (A) PPARαmRNA levels were determined by qRT-PCR and 18S ribosomal was used as a internal control (n=6 independentexperiments). (B) PPARα protein levels analyzed by Western blot (n=6 independent experiments). Data are presented asmean ± SEM. * P < 0.01.
0
0.5
1
1.5
2
2.5
ControlsiRNA
ARNTsiRNA
A
B
PPA
R-α
mR
NA
leve
ls
(fold
cha
nge)
*

B C
0
0.2
0.4
0.6
0.8
1
1.2
1.4 IgGARNT
Fold
enr
ichm
ent
18S β-actin0
0.5
1
1.5
2
2.5 IgGARNT
Fold
enr
ichm
ent
CBWD1 ELMO2 UGP2
*
* *
Supplemental Figure 9. Effective knockdown of ARNT in HEK293 cells and Positive and negative controls for ChIP studies.(A) Western blot of HEK293 cells treated with control or ARNT siRNA and blotted with ARNT antibody. (B) Negative controls forthe ChIP studies. The amplified regions do not contain any predicted HRE sites, and therefore is not regulated by ARNT (n=3).(C) Positive controls for the ChIP studies (n=3). These genes have been shown to be regulated by ARNT as described in theMethods section. CBWD1, Cobalamin Synthetase W Domain-Containing Protein 1; ELMO2, Engulfment And Cell Motility 2;UGP2, UDP-Glucose Pyrophosphorylase 2. Data are represented as fold enrichment over IgG samples and are shown asmean ± SEM. * P < 0.01 vs control. N=3 independent experiments for B and C.
ARNT
GAPDH
Control siRNA ARNT siRNAA

0
0.2
0.4
0.6
0.8
1
1.2
0
0.2
0.4
0.6
0.8
1
1.2
*
*
siRNA: Control
AR
NT
mR
NA
(fold
cha
nge)
siRNA: Control ARNT
HIF2α
HIF
2αm
RN
A (fo
ld c
hang
e)
Supplemental Figure 10. Effective reduction in the mRNA levels of ARNT (A) and its partners, HIF1α (B), HIF2α (C), and AHR(D) using siRNA approach in NRCM (n=6 independent experiments). Data are shown as mean ± SEM. * P < 0.01 vs control.
HIF
1αm
RN
A (fo
ld c
hang
e)
siRNA:
0
0.2
0.4
0.6
0.8
1
1.2
Control AHR
AH
R m
RN
A (fo
ld c
hang
e)siRNA:
*
0
0.2
0.4
0.6
0.8
1
1.2
Control HIF1α
*
A B
C D

Supplemental Figure 11. Regulation of PPARα promoter by ARNT is not mediated by HDACS. (A) PPARα mRNA levels inNRCM treated with HDAC inhibitor, trichostatin A (TSA) and in the presence and absence of ARNT siRNA. Positive controlfor TSA treatment in NRCMs treated with control-siRNA (B) or ARNT-siRNA (C). The expression of these genes is shown tobe increased with HDAC inhibition (Circ Res. 2005;97:210-218). ITPR3=inositol 1,4,5-trisphosphate receptor, type 3;JAG2=jagged 2. * P < 0.05 vs control. N=6 independent experiments for all of the panels.
* N.S.
N.S.
00.20.40.60.8
11.21.41.61.8
2PP
AR
α m
RN
A le
vels
(F
old
Chn
ge)
DMSOTSA
Control siRNA ARNT siRNA
A
0
0.5
1
1.5
2
2.5
3
ITPR3 JAG2
Rel
ativ
e Ex
pres
sion
DMSOTSA
0
0.5
1
1.5
2
2.5
3
3.5
ITPR3 JAG2
Rel
ativ
e Ex
pres
sion
DMSOTSA
**
**
B
C

Control siRNA ARNT siRNA
HDAC1
HDAC2
Tubulin
HDAC3
HDAC4
Tubulin
Control siRNA ARNT siRNAA
*
*
*
00.5
11.5
22.5
33.5
44.5
acetylated-H3 acetylated H4
Fold
Enr
ichm
ent o
ver I
gG
control-siRNAARNT-siRNA
0123456789
acetylated-H3 acetylated H4
Fold
Enr
ichm
ent o
ver I
gG
control-siRNAARNT-siRNA
NS
C DE
B
*
*
012345678
acetylated-H3 acetylated H4
Fold
Enr
ichm
ent o
ver I
gG
control-siRNAARNT-siRNA
Supplemental Figure 12. HDAC levels and histone acetylation are not altered by ARNT siRNA. (A) HDAC1, HDAC2,HDAC3, and HDAC4 protein levels in NRCM treated with control or ARNT siRNA. A Summary bar graph is shown in Panel B.ChIP studies assessing the Levels of acetylated histone H3 and H4 on the PPARα promoter (C), and on the promoter of twoknown ARNT target genes, COBW domain containing protein (CBWD, D), and UDP-glucose pyrophosphorylase-2 (UGP2, E)in HepG2 cells treated with control or ARNT siRNA. Data are presented as fold enrichment over IgG. * P < 0.05 vs control.N=3 independent experiments for panel C-E.
0
0.2
0.4
0.6
0.8
1
1.2
1.4
HDAC 1 HDAC 2 HDAC 3 HDAC 4
Prot
ein
Expr
essi
on (F
old
Cha
nge)
control-siRNA ARNT-siRNA

Rel
ativ
e ex
pres
sion
(fold
cha
nge)
Supplemental Figure 13. PPARα mRNA levels in the hearts of control and db/db mice. Data are presented as mean ± SEM. * P < 0.01. n=6 independent experiments per group.
0
0.5
1
1.5
2
2.5
control db/db
*

0
0.5
1
1.5
2
2.5
3
3.5
4
UCP2 UCP3
controlcsARNT+/-
Control csARNT-/-
Prot
ein
expr
essi
on(fo
ld c
hang
e)UCP3
UCP2
*
*
Tubulin
csARNT-/-
Supplemental Figure 14. UCP2 and UCP3 protein levels in the heart of csARNT-/- mice. A summary bar graph of the results is provided below the Western blot gels. Data are presented as mean ± SEM. * P < 0.01 versus normal chow.

xCre, ARNTf/f PPARα-/-
Cre, ARNTf/+, PPARα+/-
Cre, ARNTf/f, PPARα+/-
Cre, ARNT f/f, PPARα-/-
Cre, ARNTf/f
Cre, ARNTf/f, PPARα+/-
x
x
A
650 bp: PPARα-/-
412 bp: WT
300 bp: ARNTf/f
200 bp: WT
440 bp: Cre324 bp: WT
B
Supplemental Figure 15. Breeding scheme for generation of csARNT-/-/PPARα-/- double knockout mice. (A) Interbreedingplan of PPARα-/- and Cre, ARNTf/f Knockout mice. (B) Double knockout mice were selected by genotyping as indicated. (C)Tissue sections were stained for PPARα (green), ARNT(red), and DAPI (nuclei in blue) in hearts from control, cARNT-/- andcsARNT-/-/PPARα-/- double knockout mice. The experiment was repeated three times.
CControl csARNT-/- PPARα-/-
csARNT-/-
PPARα-/-
AR
NT
PPA
Rα
DA
PIO
verla
y

0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
0
1
2
3
4
5
6
7
8
Supplemental Figure 16. mRNA levels of (A) atrial natriuretic peptide (ANP) and (B) brain natriuretic peptide (BNP) inthe hearts of control, csARNT-/- and csARNT-/-/PPARα-/- double knockout mice 4 weeks after tamoxifen (n=5-7independent experiments). Data are presented as means ± SEM. * P < 0.05 vs control.
AN
P m
RN
A
BN
P m
RN
A
A B
Control csARNT-/- csARNT-/-
PPARα-/- Control csARNT-/- csARNT-/-
PPARα-/-
* *

Supplemental Table 1
Echocardiographic assessment of control, csARNT-/- and csARNT-/- PPARα-/- double knockout mice performed 1, 2 and 3 weeks after tamoxifen treatment and followed with one week normal chow. End-diastolic interventricular septum thickness (IVS-d); End-diastolic posterior wall thickness (LVPW-d); left ventricular diastolic internal diameter (LVID-d); fractional shorting (%FS); Ejection fraction (%EF); cardiac output (CO) and heart rate (HR) were assessed. Animal number used in each group was indicated. Data are presented as mean ± SEM. * P < 0.01 vs control.
parameter 1 week (Tomaxifen) 2 Week (Tomxifen) 3 Week ( N Chow)
group Control csARNT-/- csARNT-/- PPARα-/-
Control csARNT-/- csARNT-/- PPARα-/-
Control csARNT-/- csARNT-/- PPARα-/-
n 7 12 12 7 10 12 7 10 12 IVS-d, mm 0.71 ± 0.02 0.69 ± 0.06 0.72 ± 0.03 0.79 ± 0.07 0.61 ± 0.03* 0.67 ± 0.04 0.77 ± 0.06 0.84 ± 0.02 0.87 ± 0.01 LVPW-d, mm
0.62 ± 0.03 0.59 ± 0.05 0.64 ± 0.02 0.58 ± 0.03 0.50 ± 0.04 0.62 ± 0.03 0.61 ± 0.02 0.67 ± 0.05 0.64 ± 0.04
LVID-d, mm 3.86 ± 0.20 3.78 ± 0.144 3.77 ± 0.144 3.56 7 ± 0.15 4.18 ± 0.138* 4.01 ± 0.138 3.81 ± 0.133 4.26 ± 0.143* 3.99 ± 0.06 %FS 26.9 ± 3.33 23.29 9 ±
2.30 27.644 ±
1.12 31.33 ± 0.88 17.62 ± 1.94* 23.51 ± 1.57* 30.31 ± 1.41 21.33 ± 2.97* 27.64 ± 1.7
%EF 52.6 ± 4.49 42.0 ± 2.87* 53.9 ± 1.8 60.24 ± 1.76 38.21 ± 3.58* 47.34 ± 2.65* 61.25 ± 2.1 43.33 ± 5.36* 53.4 ± 2.62 CO, ml/min 12.72 ± 0.83 9.51 ± 14.44* 14.44 ± 0.84 14.64 ± 9.01 9.14 ± 1.45* 11.76 ± 1.48 13.50 ± 1.33 10.01 ± 1.32 16.17 ± 16.1 HR, min 445 ± 18.61 447 ± 14.1 487 ± 16.2 480 ± 21.9 488 ± 24.5 417 ± 15.5 489 ± 11.1 415 ± 15.5 427 ± 13.5