AACR poster Yang
Transcript of AACR poster Yang

Yang Cong, Ming Zhan, Stephen T. C. Wong NCI Center for Modeling Cancer Development, Department of Systems Medicine and
Bioengineering; Houston Methodist Research Institute, Weill Cornel Medical College
[1] Mani, S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cell. Cell 133, 704-15 (2008).
[2] Tam, W. L. et al. Protein Kinase C α Is a Central Signaling Node and Therapeutic Target for Breast Cancer Stem Cells. Cancer Cell 24, 347-64 (2013).
[3] Liu, S. L. et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem Cell
Reports 2, 78-91 (2014).
Liu S. L. et al. showed that breast cancer stem cells display plasticity that enables them
to transition between epithelial-like (ALDH+) and mesenchymal-like (CD24-CD44+)
states [3].
Out computational model evaluates their breast cancer patients’ gene profiling data and
find some quasi-epithelial and quasi-mesenchymal gene expression patterns with high
stability scores. This may indicate the existence of intermediate stage in epithelial-
mesenchymal (EMT) and mesenchymal-epithelial (MET) transition.
ABSTRACT # 3998 Network-based Identification of Gene Signatures of
Epithelial-mesenchymal Transition in Breast Cancer
Method
1. Transitions in cell phenotypes from epithelial to mesenchymal states, defined as epithelial-
mesenchymal transition (EMT) contributes to tumor progression.
2. Induction of EMT in immortalized human mammary epithelial cells has shown to result in
expression of stem-cell markers and acquisition of functional stem-cell properties [1].
Background
Results
Discussion
References
Acknowledgement
This work is supported by NIH grant U54CA149196 and John S. Dunn Research Foundation..
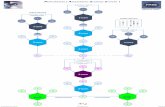
1. EMT regulatory pathway
was built up based on
Ingenuity Knowledge Base.
2. Gene expression profiles of
breast cancer patients were
extracted from The Cancer
Genome Atlas (TCGA) and
Gene Expression Omnibus
(GEO).
3. Based on gene profiling
data, we modeled the gene
regulatory mechanism of
EMT by a stochastic model.
4. An efficient computational
algorithm was developed to
identify gene signatures.
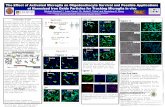
Our computational model identified and evaluates
gene expression patterns of epithelial and
mesenchymal cells.
The results are confirmed with data from Tam W. L.
et al [2]..They transduced genes encoding Snail and
Slug into immortalized human mammary epithelial
cells (HMELs), resulting EMT-TF-induced
mesenchymal cells, phase contrast images and
qPCR shown in the left.