A demonstration of clustering in protein contact maps for α-helix pairs Robert Fraser, University...
Transcript of A demonstration of clustering in protein contact maps for α-helix pairs Robert Fraser, University...

A demonstration of clustering in protein contact maps for α-helix pairs
Robert Fraser, University of Waterloo
Janice Glasgow, Queen’s University

2
Hypothesis
Properties of the three-dimensional configuration of a pair of α-helices may be predicted from the contact map corresponding to the pair.

3
What to expect
• The α-helix
• Protein structure prediction
• Packing of helices
• Prediction of packing
• Future work

4
Where do proteins come from?
What we’re interested in
Image from http://www.accessexcellence.org/RC/VL/GG/images/protein_synthesis.gif

5
Amino acid residues
• Two representations of the same α-helix
• Left helix shows residues only
• Right helix shows all backbone atoms

6
Why α-helices?
• Protein Structure Prediction– The 3D structure of a
protein is primarily determined by groups of secondary structures
– The α-helix is the most common secondary structure

7
Protein Structure Prediction
• is hard.
• There are many different approaches• Crystallization techniques are time-consuming• Tryptych
– Case based-reasoning approach to prediction– Uses different experts as advisors– Works from a contact map

8
The Goal
• 2D representation 3D• Toy example:
A C
BC 4 5 0
B 3 0 5
A 0 3 4
A B C

9
The Goal
If we apply a threshold of 4.5 units, can we still determine the shape?
C 4 5 0
B 3 0 5
A 0 3 4
A B C
A C
B
C T F T
B T T F
A T T T
A B C
?

10
Protein Structure Prediction• Build (or predict) contact map• Predict the three-dimensional structure from the
contact map• N×N matrix, N= # of amino acids in the protein• C(i,j) = true if the distance from amino acid i to j is
less than a threshold (ie. 7Å)
?

11
Refining a contact map
Contact map for entire protein
Contact map for pair of α-helices
Interface region of contact map

12
Hypothesis (again)
Properties of the three-dimensional configuration of a pair of α-helices may be predicted from the contact map corresponding to the pair.

13
Packing of helices
• A nice simple boolean property
• Found by rotating both helices to be axis-aligned, then comparing coordinates
packing non-packing
90°not 90°

14
Prediction of packing
Need a map comparison method

15
Comparing maps
• Comparing two maps is not straightforward
• Reflections are insignificant

16
Comparing maps
• Translations are (generally) insignificant

17
Prediction of packing
• Classify the maps based on contact locations
central corneredge

18
Packing classes

19
Packing data by class
• No clustering to be done on the central and edge classes
• Reflections of each corner map were used to build the data set

20
Clustering Corner Maps
• Each corner map is transformed to a 15×15 map• A map corresponds to a 225 character binary string• Distance can measure using cosine

21
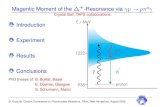
Clustering Results

22
Nearest Neighbour
• Closest neighbour by cosine distance
• Of the 862 contact maps in the data set, only 9 had a packing value different from the nearest neighbour
• Almost 99% accuracy!

23
Contributions
• Prediction of helix pair properties– 1st to establish that contact maps cluster– Developed a novel contact map classification
scheme for helix pairs– 1st to demonstrate that helix pair properties
may be predicted from contact maps

24
Future Work
• Implement the packing advisor (done)
• Explore other properties of α-helices that could be predicted from contact maps– Interhelical angle– Interhelical distance

25
Thanks
• Questions?
Supported by
NSERC, PRECARN IRIS, Queen’s

26
Clustering Results
Non-packing Mixed
Packing

27
Levels of Structure
Primary
Secondary

28
α-helix properties

29
Tryptych

30
Chothia packing model